













| General Information: |
|
| Name(s) found: |
UGPA1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 470 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA]
|
| Biological Process: |
response to salt stress
[IEP]
metabolic process [ISS] response to cadmium ion [IEP] |
| Molecular Function: |
nucleotidyltransferase activity
[IEA]
UTP:glucose-1-phosphate uridylyltransferase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAATATEKLP QLKSAVDGLT EMSENEKSGF INLVSRYLSG EAQHIEWSKI QTPTDEIVVP 60
61 YDKMANVSED ASETKYLLDK LVVLKLNGGL GTTMGCTGPK SVIEVRDGLT FLDLIVIQIE 120
121 NLNNKYNCKV PLVLMNSFNT HDDTQKIVEK YTKSNVDIHT FNQSKYPRVV ADEFVPWPSK 180
181 GKTDKDGWYP PGHGDVFPSL MNSGKLDAFL SQGKEYVFIA NSDNLGAIVD LKILKHLIQN 240
241 KNEYCMEVTP KTLADVKGGT LISYEGKVQL LEIAQVPDEH VNEFKSIEKF KIFNTNNLWV 300
301 NLKAIKKLVE ADALKMEIIP NPKEVDGVKV LQLETAAGAA IRFFDNAIGV NVPRSRFLPV 360
361 KATSDLLLVQ SDLYTLVDGF VTRNKARTNP TNPAIELGPE FKKVASFLSR FKSIPSIVEL 420
421 DSLKVSGDVW FGSGVVLKGK VTVKANAGTK LEIPDNAVLE NKDINGPEDL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
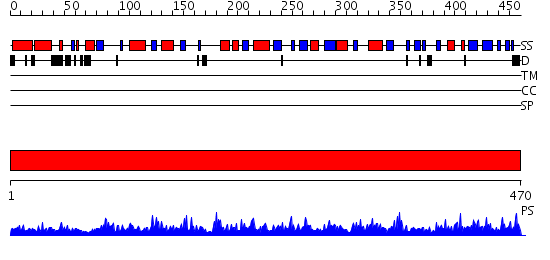
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..470] | 1000.0 | X-ray structure of gene product from arabidopsis thaliana at3g03250, a putative UDP-glucose pyrophosphorylase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)