













| General Information: |
|
| Name(s) found: |
UAP2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 502 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
metabolic process
[IEA][ISS]
|
| Molecular Function: |
nucleotidyltransferase activity
[IEA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKEPTTEIEI ETSAVATILP PPLPPTASPH QALVERLKDY GQEDVFSLWD ELSPEERDLL 60
61 LRDIENLDLP RIDRIIRCSL HSQGLPVAAI EPVPENCVST VEERTKEDRE KWWKMGLKAI 120
121 YEGKLGVVLL SGGQGTRLGS SDPKGCYNIG LPSGKSLFQI QAERILCVQR LASQAMSEAS 180
181 PTRPVTIQWY IMTSPFTHEP TQKFFKSHKY FGLEPDQVTF FQQGTLPCIS KDGKFIMETP 240
241 FSLSKAPDGN GGVYTALKSS RLLEDMASRG IKYVDCYGVD NVLVRVADPT FLGYFIDKSA 300
301 ASAAKVVRKA YPQEKVGVFV RRGKGGPLTV VEYTELDQSM ASATNQQTGR LQYCWSNVCL 360
361 HMFTLDFLNQ VANGLEKDSV YHLAEKKIPS INGDIVGLKL EQFIFDCFPY APSTALFEVL 420
421 REEEFAPVKN ANGSNYDTPE SARLLVLRLH TRWVIAAGGF LTHSVPLYAT GVEVSPLCSY 480
481 AGENLEAICR GRTFHAPCEI SL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
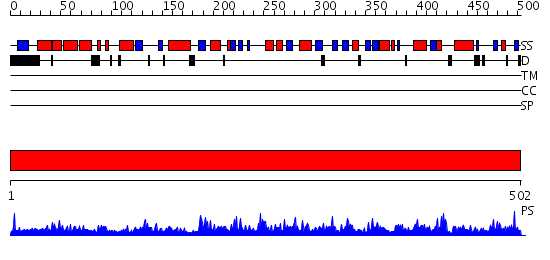
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..502] | 165.0 | Crystal structure of UDP-N-acetylglucosamine pyrophosphorylase (Agx2) from Mus musculus at 2.50 A resolution |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)