













| General Information: |
|
| Name(s) found: |
SUVH4_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 624 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
peptidyl-lysine methylation
[IDA]
histone methylation [IDA] maintenance of DNA methylation [IDA] histone H3-K9 methylation [IMP] |
| Molecular Function: |
double-stranded methylated DNA binding
[IDA]
methyl-CpNpG binding [IDA] methyl-CpNpN binding [IDA] histone methyltransferase activity (H3-K9 specific) [IDA] methyl-CpG binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAGKRKRANA PDQTERRSSV RVQKVRQKAL DEKARLVQER VKLLSDRKSE ICVDDTELHE 60
61 KEEENVDGSP KRRSPPKLTA MQKGKQKLSV SLNGKDVNLE PHLKVTKCLR LFNKQYLLCV 120
121 QAKLSRPDLK GVTEMIKAKA ILYPRKIIGD LPGIDVGHRF FSRAEMCAVG FHNHWLNGID 180
181 YMSMEYEKEY SNYKLPLAVS IVMSGQYEDD LDNADTVTYT GQGGHNLTGN KRQIKDQLLE 240
241 RGNLALKHCC EYNVPVRVTR GHNCKSSYTK RVYTYDGLYK VEKFWAQKGV SGFTVYKYRL 300
301 KRLEGQPELT TDQVNFVAGR IPTSTSEIEG LVCEDISGGL EFKGIPATNR VDDSPVSPTS 360
361 GFTYIKSLII EPNVIIPKSS TGCNCRGSCT DSKKCACAKL NGGNFPYVDL NDGRLIESRD 420
421 VVFECGPHCG CGPKCVNRTS QKRLRFNLEV FRSAKKGWAV RSWEYIPAGS PVCEYIGVVR 480
481 RTADVDTISD NEYIFEIDCQ QTMQGLGGRQ RRLRDVAVPM NNGVSQSSED ENAPEFCIDA 540
541 GSTGNFARFI NHSCEPNLFV QCVLSSHQDI RLARVVLFAA DNISPMQELT YDYGYALDSV 600
601 HGPDGKVKQL ACYCGALNCR KRLY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
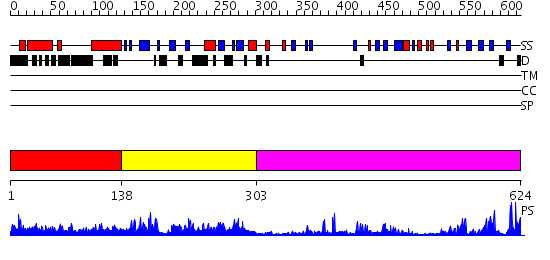
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..137] | 7.69897 | No description for 2e6sA was found. | |
| 2 | View Details | [138..302] | 46.886057 | No description for PF02182.8 was found. No confident structure predictions are available. | |
| 3 | View Details | [303..624] | 47.045757 | No description for 2igqA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)