













| General Information: |
|
| Name(s) found: |
SSRP1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 646 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
FACT complex [IDA] nuclear euchromatin [IDA] |
| Biological Process: | NONE FOUND |
| Molecular Function: |
transcription factor activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MADGHSFNNI SLSGRGGKNP GLLKINSGGI QWKKQGGGKA VEVDRSDIVS VSWTKVTKSN 60
61 QLGVKTKDGL YYKFVGFRDQ DVPSLSSFFQ SSYGKTPDEK QLSVSGRNWG EVDLHGNTLT 120
121 FLVGSKQAFE VSLADVSQTQ LQGKNDVTLE FHVDDTAGAN EKDSLMEISF HIPNSNTQFV 180
181 GDENRPPSQV FNDTIVAMAD VSPGVEDAVV TFESIAILTP RGRYNVELHL SFLRLQGQAN 240
241 DFKIQYSSVV RLFLLPKSNQ PHTFVVISLD PPIRKGQTMY PHIVMQFETD TVVESELSIS 300
301 DELMNTKFKD KLERSYKGLI HEVFTTVLRW LSGAKITKPG KFRSSQDGFA VKSSLKAEDG 360
361 VLYPLEKGFF FLPKPPTLIL HDEIDYVEFE RHAAGGANMH YFDLLIRLKT DHEHLFRNIQ 420
421 RNEYHNLYTF ISSKGLKIMN LGGAGTADGV AAVLGDNDDD DAVDPHLTRI RNQAADESDE 480
481 EDEDFVMGED DDGGSPTDDS GGDDSDASEG GVGEIKEKSI KKEPKKEASS SKGLPPKRKT 540
541 VAADEGSSKR KKPKKKKDPN APKRAMSGFM FFSQMERDNI KKEHPGIAFG EVGKVLGDKW 600
601 RQMSADDKEP YEAKAQVDKQ RYKDEISDYK NPQPMNVDSG NDSDSN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
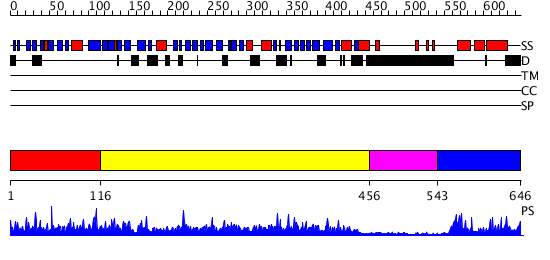
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..115] | 2.042969 | View MSA. Confident ab initio structure predictions are available. | |
| 2 | View Details | [116..455] | 93.522879 | No description for 2gcjA was found. | |
| 3 | View Details | [456..542] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [543..646] | 25.69897 | TERNARY COMPLEX OF THE DNA BINDING DOMAINS OF THE OCT1 AND SOX2 TRANSCRIPTION FACTORS WITH A 19MER OLIGONUCLEOTIDE FROM THE HOXB1 REGULATORY ELEMENT |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)