













| General Information: |
|
| Name(s) found: |
PARP2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 637 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
protein amino acid ADP-ribosylation
[IEA][ISS]
|
| Molecular Function: |
nucleic acid binding
[IEA]
NAD+ ADP-ribosyltransferase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MANKLKVDEL RLKLAERGLS TTGVKAVLVE RLEEAIAEDT KKEESKSKRK RNSSNDTYES 60
61 NKLIAIGEFR GMIVKELREE AIKRGLDTTG TKKDLLERLC NDANNVSNAP VKSSNGTDEA 120
121 EDDNNGFEEE KKEEKIVTAT KKGAAVLDQW IPDEIKSQYH VLQRGDDVYD AILNQTNVRD 180
181 NNNKFFVLQV LESDSKKTYM VYTRWGRVGV KGQSKLDGPY DSWDRAIEIF TNKFNDKTKN 240
241 YWSDRKEFIP HPKSYTWLEM DYGKEENDSP VNNDIPSSSS EVKPEQSKLD TRVAKFISLI 300
301 CNVSMMAQHM MEIGYNANKL PLGKISKSTI SKGYEVLKRI SEVIDRYDRT RLEELSGEFY 360
361 TVIPHDFGFK KMSQFVIDTP QKLKQKIEMV EALGEIELAT KLLSVDPGLQ DDPLYYHYQQ 420
421 LNCGLTPVGN DSEEFSMVAN YMENTHAKTH SGYTVEIAQL FRASRAVEAD RFQQFSSSKN 480
481 RMLLWHGSRL TNWAGILSQG LRIAPPEAPV TGYMFGKGVY FADMFSKSAN YCYANTGAND 540
541 GVLLLCEVAL GDMNELLYSD YNADNLPPGK LSTKGVGKTA PNPSEAQTLE DGVVVPLGKP 600
601 VERSCSKGML LYNEYIVYNV EQIKMRYVIQ VKFNYKH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
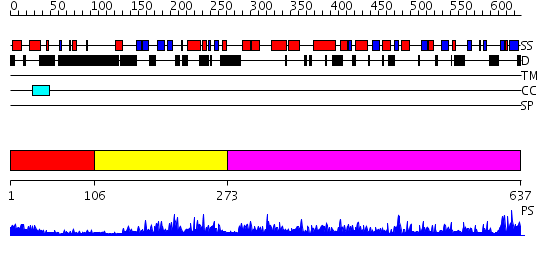
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..105] | 7.154902 | Solution structure of BRCT domain of poly(ADP-ribose) polymerase-1 | |
| 2 | View Details | [106..272] | 36.69897 | Solution structure of WGR domain of poly(ADP-ribose) polymerase-1 | |
| 3 | View Details | [273..637] | 124.0 | Crystal structure of catalytic domain of human poly(ADP-ribose) polymerase with a novel inhibitor |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)