













| General Information: |
|
| Name(s) found: |
PMG1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 557 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
cytosol [IDA] mitochondrial envelope [IDA] plasma membrane [IDA] |
| Biological Process: |
response to cold
[IEP]
response to cadmium ion [IEP] |
| Molecular Function: |
metal ion binding
[IEA]
catalytic activity [IEA] phosphoglycerate mutase activity [IEA] manganese ion binding [IEA] 2,3-bisphosphoglycerate-independent phosphoglycerate mutase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATSSAWKLD DHPKLPKGKT IAVIVLDGWG ESAPDQYNCI HNAPTPAMDS LKHGAPDTWT 60
61 LIKAHGTAVG LPSEDDMGNS EVGHNALGAG RIFAQGAKLC DQALASGKIF EGEGFKYVSE 120
121 SFETNTLHLV GLLSDGGVHS RLDQLQLLIK GSAERGAKRI RVHILTDGRD VLDGSSVGFV 180
181 ETLEADLVAL RENGVDAQIA SGGGRMYVTL DRYENDWEVV KRGWDAQVLG EAPHKFKNAV 240
241 EAVKTLRKEP GANDQYLPPF VIVDESGKAV GPIVDGDAVV TFNFRADRMV MHAKALEYED 300
301 FDKFDRVRYP KIRYAGMLQY DGELKLPSRY LVSPPEIDRT SGEYLTHNGV STFACSETVK 360
361 FGHVTFFWNG NRSGYFNEKL EEYVEIPSDS GISFNVQPKM KALEIGEKAR DAILSGKFDQ 420
421 VRVNIPNGDM VGHTGDIEAT VVACEAADLA VKMIFDAIEQ VKGIYVVTAD HGNAEDMVKR 480
481 DKSGKPALDK EGKLQILTSH TLKPVPIAIG GPGLAQGVRF RKDLETPGLA NVAATVMNLH 540
541 GFVAPSDYEP TLIEVVE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
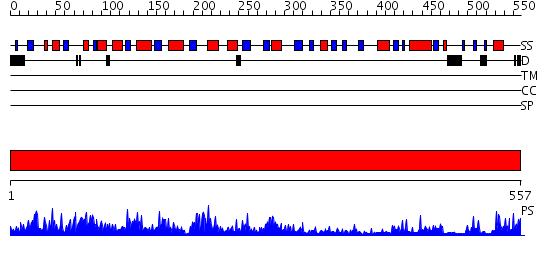
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..557] | 180.0 | 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, substrate-binding domain; 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, catalytic domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)