













| General Information: |
|
| Name(s) found: |
PEM2_ARATH
[Swiss-Prot]
PEAM2_ARATH [Swiss-Prot] |
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 475 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
metabolic process
[IEA]
|
| Molecular Function: |
methyltransferase activity
[IEA]
phosphoethanolamine N-methyltransferase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEHSSDLTVE AMMLDSKASD LDKEERPEVL SLIPPYEGKS VLELGAGIGR FTGELAQKAG 60
61 EVIALDFIES AIQKNESVNG HYKNIKFMCA DVTSPDLKIK DGSIDLIFSN WLLMYLSDKE 120
121 VELMAERMIG WVKPGGYIFF RESCFHQSGD SKRKSNPTHY REPRFYTKVF QECQTRDASG 180
181 NSFELSMVGC KCIGAYVKNK KNQNQICWIW QKVSVENDKD FQRFLDNVQY KSSGILRYER 240
241 VFGEGYVSTG GFETTKEFVA KMDLKPGQKV LDVGCGIGGG DFYMAENFDV HVVGIDLSVN 300
301 MISFALERAI GLKCSVEFEV ADCTTKTYPD NSFDVIYSRD TILHIQDKPA LFRTFFKWLK 360
361 PGGKVLITDY CRSAETPSPE FAEYIKQRGY DLHDVQAYGQ MLKDAGFDDV IAEDRTDQFV 420
421 QVLRRELEKV EKEKEEFISD FSEEDYNDIV GGWSAKLERT ASGEQKWGLF IADKK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
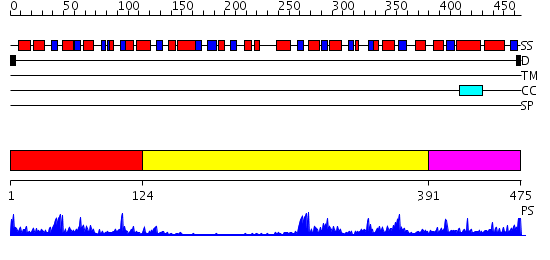
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..123] | 13.09691 | No description for 2i6gA was found. | |
| 2 | View Details | [124..390] | 8.522879 | The crystal structure of E. coli Fmu binary complex with S-Adenosylmethionine at 2.1 A resolution | |
| 3 | View Details | [391..475] | 41.30103 | CmaA1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 |
|
|||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)