













| General Information: |
|
| Name(s) found: |
PLDA3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 820 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IEA]
|
| Biological Process: |
response to abscisic acid stimulus
[IMP]
response to salt stress [IMP] membrane lipid catabolic process [IMP] response to water deprivation [IMP] |
| Molecular Function: |
phospholipase D activity
[IDA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTEQLLLHGT LEVKIYRIDK LHQRSRFNLC GKGNKEPTGK KTQSQIKRLT DSCTSLFGGH 60
61 LYATIDLDRS RVARTMMRRH PKWLQSFHVY TAHSISKIIF TVKEDEPVSA SLIGRAYLPV 120
121 TEVITGQPID RWLDILDENR RPIQGGSKLH VRVKFTHVTQ DVNWNKGIIL PSFNGVPNAY 180
181 FNQREGCKVT LYQDAHVLNE YPDVTLTGGQ VIYKHHRCWE EIFDAIWEAK HLIYIAGWSV 240
241 NTDVTLVRDP KRTRPGGDLK LGELLKKKAE ENVTVLMLVW DDRTSHEVFK RDGLMMTHDQ 300
301 ETYDYFKNTK VRCVLCPRNP DNGDSIVQGF EVATMFTHHQ KTIVVDSEVD GSLTKRRIVS 360
361 FLGGIDLCDG RYDTVEHPLF GTLNSVHAND FHQPNFDGAS IKKGGPREPW HDIHCKLDGP 420
421 AAWDVLYNFE QRWMKQGSGR RYLISMAQLA EITVPPLPIV QPDNEEGWTV QVFRSIDDGA 480
481 VEGFPEDPRE AASIGLISGK DNVIERSIQD AYVNAIRRAK NFIYIENQYF LGSSFGWNSR 540
541 DINLNEINAL QLIPKEISLK IVSKIEAGER FSVYIVIPLW PEGKPGSASV QAILDWQRRT 600
601 MEMMYTDIII ALRKKGLDAN PRDYLTFFCL GNREKGKVGE YLPPEKPEAN SDYARAQESR 660
661 RFMIYVHSKM MIVDDEYIII GSANINQRSM DGGRDTEIAM GAYQPSHLLS TNNMRPVGQI 720
721 FSFRISLWLE HLRVTTNAFQ CPESEECIRM VNATADELWG LYSAQEYPRN DDLPGHLLSY 780
781 PISIGSNGEV TNLAGTEFFP DTNAKVVGEK SNYLPPILTS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
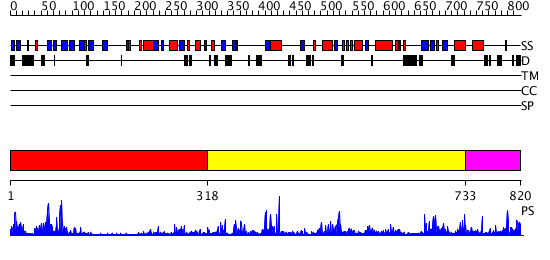
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..317] | 25.69897 | Synaptogamin I | |
| 2 | View Details | [318..732] | 5.39794 | No description for 2ze4A was found. | |
| 3 | View Details | [733..820] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)