













| General Information: |
|
| Name(s) found: |
PLDG3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 866 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IEA]
|
| Biological Process: |
phosphatidylcholine metabolic process
[IEA]
metabolic process [IEA] |
| Molecular Function: |
phospholipase D activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAYHPVYNET MSMGGGSSNE FGQWLDKQLV PFDTSSGSLR VELLHGNLDI WVKEAKHLPN 60
61 MDGFHNTLVG GMFFGLGRRN HKVDGENSSK ITSDPYVTVS ISGAVIGRTF VISNSENPVW 120
121 MQHFDVPVAH SAAKVHFVVK DSDIIGSQII GAVEIPTEQL CSGNRIEGLF PILNSRGKPC 180
181 KQGAVLSLSI QYIPMERMRL YQKGVGFGVE CVGVPGTYFP LRKGGRVTLY QDAHVDDGTL 240
241 PSVHLDGGIQ YRHGKCWEDM ADAIRRARRL IYITGWSVFH PVRLVRRNND PTQGTLGELL 300
301 KVKSQEGVRV LVLVWDDPTS RSLLGFSTKG LMNTSDEETR RFFKHSSVQV LLCPRYGGKG 360
361 HSFIKKSEVE TIYTHHQKTM IVDAEAAQNR RKIVAFVGGL DLCNGRFDTP KHPLFRTLKT 420
421 IHKDDFHNPN FVTTADDGPR EPWHDLHSKI DGPAAYDVLA NFEERWMKAS KPRGIGRLRT 480
481 SSDDSLLRLD RIPDIMGLSE ASSANDNDPE SWHVQVFRSI DSSSVKGFPK DPKEATGRNL 540
541 LCGKNILIDM SIHAAYVKAI RSAQHFIYIE NQYFLGSSFN WDSNKNLGAN NLIPMEIALK 600
601 IANKIRAREK FAAYIVIPMW PEGAPTSNPI QRILYWQHKT MQMMYQTIYK ALVEVGLDGQ 660
661 LEPQDFLNFF CLGTREVGTR EVPDGTVSVY NSPRKPPQLN AAQVQALKSR RFMIYVHSKG 720
721 MVVDDEFVLI GSANINQRSL EGTRDTEIAM GGYQPHHSWA KKGSRPRGQI FGYRMSLWAE 780
781 HLGFLEQEFE EPENMECVRR VRQLSELNWR QYAAEEVTEM PGHLLKYPVQ VDRTGKVSSL 840
841 PGYETFPDLG GKIIGSFLVV EENLTI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
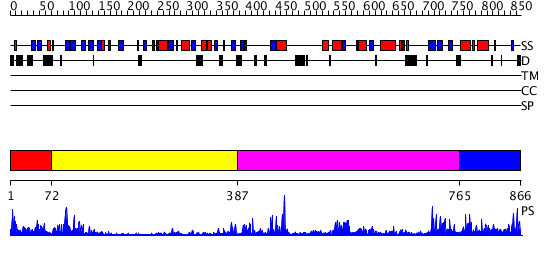
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..71] | 30.30103 | Synaptogamin I | |
| 2 | View Details | [72..386] | 9.39794 | Domain from cytosolic phospholipase A2; Cytosolic phospholipase A2 catalytic domain | |
| 3 | View Details | [387..764] | 13.0 | Phospholipase D | |
| 4 | View Details | [765..866] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)