













| General Information: |
|
| Name(s) found: |
PLDG1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 858 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA]
|
| Biological Process: |
defense response to bacterium, incompatible interaction
[IGI]
embryonic development ending in seed dormancy [IMP] |
| Molecular Function: |
protein binding
[IPI]
phospholipase D activity [IDA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAYHPAYTET MSMGGGSSHG GGQQYVPFAT SSGSLRVELL HGNLDIWVKE AKHLPNMDGF 60
61 HNRLGGMLSG LGRKKVEGEK SSKITSDPYV TVSISGAVIG RTFVISNSEN PVWMQHFDVP 120
121 VAHSAAEVHF VVKDSDIIGS QIMGAVGIPT EQLCSGNRIE GLFPILNSSG KPCKQGAVLG 180
181 LSIQYTPMER MRLYQMGVGS GNECVGVPGT YFPLRKGGRV TLYQDAHVDD GTLPSVHLDG 240
241 GIQYRHGKCW EDMADAIRQA RRLIYITGWS VFHPVRLVRR TNDPTEGTLG ELLKVKSQEG 300
301 VRVLVLVWDD PTSRSLLGFK TQGVMNTSDE ETRRFFKHSS VQVLLCPRSG GKGHSFIKKS 360
361 EVGTIYTHHQ KTVIVDAEAA QNRRKIVAFV GGLDLCNGRF DTPKHPLFRT LKTLHKDDFH 420
421 NPNFVTTADD GPREPWHDLH SKIDGPAAYD VLANFEERWM KASKPRGIGK LKSSSDDSLL 480
481 RIDRIPDIVG LSEASSANDN DPESWHVQVF RSIDSSSVKG FPKDPKEATG RNLLCGKNIL 540
541 IDMSIHAAYV KAIRSAQHFI YIENQYFLGS SFNWDSNKDL GANNLIPMEI ALKIANKIRA 600
601 REKFAAYIVI PMWPEGAPTS NPIQRILYWQ HKTMQMMYQT IYKALVEVGL DSQFEPQDFL 660
661 NFFCLGTREV PVGTVSVYNS PRKPPQPNAN ANAAQVQALK SRRFMIYVHS KGMVVDDEFV 720
721 LIGSANINQR SLEGTRDTEI AMGGYQPHYS WAMKGSRPHG QIFGYRMSLW AEHLGFLEQG 780
781 FEEPENMECV RRVRQLSELN WRQYAAEEVT EMSGHLLKYP VQVDRTGKVS SLPGCETFPD 840
841 LGGKIIGSFL ALQENLTI |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
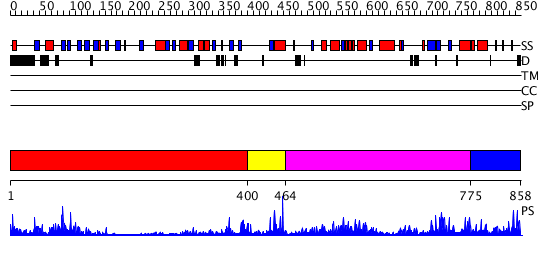
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..399] | 11.30103 | Domain from cytosolic phospholipase A2; Cytosolic phospholipase A2 catalytic domain | |
| 2 | View Details | [400..463] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [464..774] | 4.0 | Crystal Structure of Escherichia coli Polyphosphate Kinase | |
| 4 | View Details | [775..858] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)