













| General Information: |
|
| Name(s) found: |
PLDA4_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 762 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA]
|
| Biological Process: |
root development
[IMP]
cell division [IMP] cellular response to potassium ion starvation [IMP] positive regulation of nitrogen utilization [IMP] phospholipid catabolic process [IDA][TAS] post-embryonic development [IMP] response to osmotic stress [IMP] cellular response to phosphate starvation [IMP] cell growth [IMP] cellular response to nitrogen starvation [IMP] |
| Molecular Function: |
phospholipase D activity
[IDA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MELEEQKKYF HGTLEITIFD ATPFSPPFPF NCICTKPKAA YVTIKINKKK VAKTSSEYDR 60
61 IWNQTFQILC AHPVTDTTIT ITLKTRCSVL GRFRISAEQI LTSNSAVING FFPLIADNGS 120
121 TKRNLKLKCL MWFRPAYLEP GWCRALEEAS FQGIRNASFP QRSNCRVVLY QDAHHKATFD 180
181 PRVDDVPFNA RNLWEDVYKA IESARHLVYI AGWALNPNLV LVRDNETEIP HAVGVTVGEL 240
241 LKRKSEEGVA VRVMLWNDET SLPMIKNKGV MRTNVERALA YFRNTNVVCR LCPRLHKKLP 300
301 TAFAHHQKTI TLDTRVTNSS TKEREIMSFL GGFDLCDGRY DTEEHSLFRT LGTEADFYQT 360
361 SVAGAKLSRG GPREPWHDCH VSVVGGAAWD VLKNFEQRWT KQCNPSVLVN TSGIRNLVNL 420
421 TGPTEENNRK WNVQVLRSID HISATEMPRG LPVEKSVHDG YVAAIRKAER FIYIENQYFM 480
481 GSCDHWESKN DKICSGCTNL IPVEIALKIA AKIRARERFA VYIVIPMWPE GPPESETVEE 540
541 ILHWTRETMS MMYQIIGEAI WEVGDKSHPR DYLNFFCLAN REEKRDGEFE AVSSPHQKTH 600
601 YWNAQRNRRF MVYVHSKLMI VDDTYILIGS ANINQRSMDG CRDTEIAIGC YQTNTNNTNE 660
661 IQAYRLSLWY EHTGGKITAD DLSSSEPESL ECVRGLRTIG EQMWEIYSGD KVVDMLGIHL 720
721 VAYPISVTGD GAVEEVGDGC FPDTKTLVKG KRSKMFPPVL TT |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
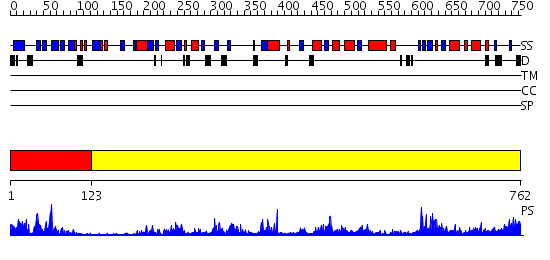
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..122] | 13.522879 | Crystallographic Identification of Sr2+ Coordination Site in Synaptotagmin I C2B Domain | |
| 2 | View Details | [123..762] | 6.62 | Tungstate-inhibited phospholipase D from Streptomyces sp. strain PMF. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)