













| General Information: |
|
| Name(s) found: |
PLDA2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 810 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast envelope
[IDA]
|
| Biological Process: |
phosphatidylcholine metabolic process
[IEA]
metabolic process [IEA] |
| Molecular Function: |
phospholipase D activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEECLLHGRL HATIYEVDHL HAEGGRSGFL GSILANVEET IGVGKGETQL YATIDLEKAR 60
61 VGRTRKITKE PKNPKWFESF HIYCGHMAKH VIFTVKDANP IGATLIGRGY IPVEDILHGE 120
121 EVDRWVDILD NEKNPIAGGS KIHVKLQYFG VEKDKNWNRG IKSAKFPGVP YTFFSQRRGC 180
181 KVSLYQDAHI PGNFVPKIPL AGGKNYEPHR CWEDIFDAIT NAKHLIYITG WSVYTEISLV 240
241 RDSRRPKQGG DVTVGELLKK KASEGVKVIL LVWDDRTSVD LLKKDGLMAT HDEETENFFR 300
301 GTDVNCILCP RNPDDGGSIV QNLQISTMFT HHQKIVVVDS EMPSGGSRSR RIVSFVGGLD 360
361 LCDGRYDTPF HSLFRTLDTA HHDDFHQPNF TGAAITKGGP REPWHDIHCR LEGPIAWDVL 420
421 YNFEQRWSRQ GGKDILVKMR ELGDIIIPPS PVLFSEDHDV WNVQLFRSID GGAAAGFPDS 480
481 PEAAAEAGLV SGKDNIIDRS IQDAYIHAIR RAKDFIYIEN QYFLGSSFAW SADGIKPEEI 540
541 NALHLIPKEL SLKIVSKIKA GEKFKVYVVV PMWPEGIPES GSVQAILDWQ KRTMEMMYKD 600
601 VIKALRENGL EGEDPRDYLT FFCLGNREVK KDGEYEPSEK PEPDTDYIRA QEARRFMIYV 660
661 HTKMMIVDDE YIIIGSANIN QRSMDGARDS EIAMGGYQPY HLSTRQPARG QIHGFRMSLW 720
721 YEHLGMLDET FLDPSSQECI QKVNRVADKY WDLYSSESLE HDLPGHLLRY PIGIASEGNI 780
781 TELPGCEFFP DTKARILGVK SDYMPPILTT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
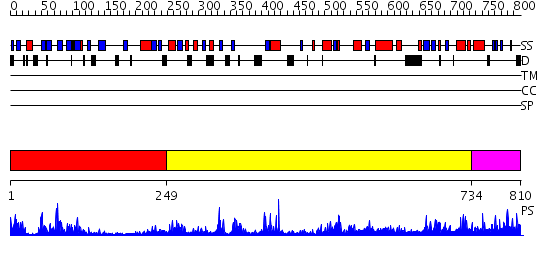
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..248] | 24.69897 | Synaptogamin I | |
| 2 | View Details | [249..733] | 17.69897 | Phospholipase D | |
| 3 | View Details | [734..810] | 1.118981 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 |
|
||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)