













| General Information: |
|
| Name(s) found: |
NIA1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 917 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[TAS]
|
| Biological Process: |
nitric oxide biosynthetic process
[IMP]
response to light stimulus [IMP] nitrate assimilation [IMP] |
| Molecular Function: |
protein binding
[IPI]
nitrate reductase activity [IMP] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATSVDNRHY PTMNGVAHAF KPPLVPSPRS FDRHRHQNQT LDVILTETKI VKETEVITTV 60
61 VDSYDDSSSD DEDESHNRNV PYYKELVKKS NSDLEPSILD PRDESTADSW IQRNSSMLRL 120
121 TGKHPFNAEA PLPRLMHHGF ITPVPLHYVR NHGAVPKANW SDWSIEITGL VKRPAKFTME 180
181 ELISEFPSRE FPVTLVCAGN RRKEQNMVKQ TIGFNWGSAG VSTSLWKGIP LSEILRRCGI 240
241 YSRRGGALNV CFEGAEDLPG GGGSKYGTSI KKEMAMDPAR DIILAYMQNG ELLTPDHGFP 300
301 VRVIVPGFIG GRMVKWLKRI IVTPQESDSY YHYKDNRVLP SLVDAELANS EAWWYKPEYI 360
361 INELNINSVI TTPGHAEILP INAFTTQKPY TLKGYAYSGG GKKVTRVEVT LDGGDTWSVC 420
421 ELDHQEKPNK YGKFWCWCFW SLDVEVLDLL SAKDVAVRAW DESFNTQPDK LIWNLMGMMN 480
481 NCWFRIRTNV CKPHRGEIGI VFEHPTRPGN QSGGWMAKER QLEISSESNN TLKKSVSSPF 540
541 MNTASKMYSI SEVRKHNTAD SAWIIVHGHI YDCTRFLKDH PGGTDSILIN AGTDCTEEFE 600
601 AIHSDKAKKL LEDYRIGELI TTGYDSSPNV SVHGASNFGP LLAPIKELTP QKNIALVNPR 660
661 EKIPVRLIEK TSISHDVRKF RFALPSEDQQ LGLPVGKHVF VCANINDKLC LRAYTPTSAI 720
721 DAVGHIDLVV KVYFKDVHPR FPNGGLMSQH LDSLPIGSMI DIKGPLGHIE YKGKGNFLVS 780
781 GKPKFAKKLA MLAGGTGITP IYQIIQSILS DPEDETEMYV VYANRTEDDI LVREELEGWA 840
841 SKHKERLKIW YVVEIAKEGW SYSTGFITEA VLREHIPEGL EGESLALACG PPPMIQFALQ 900
901 PNLEKMGYNV KEDLLIF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
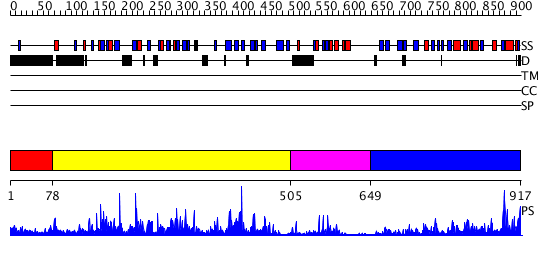
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..77] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [78..504] | 108.0 | crystal structure of the Molybdenum-containing nitrate reducing fragment of Pichia angusta assimilatory nitrate reductase | |
| 3 | View Details | [505..648] | 8.30103 | L230A mutant flavocytochrome b2 with benzoylformate | |
| 4 | View Details | [649..917] | 64.0 | STRUCTURAL STUDIES ON CORN NITRATE REDUCTASE: REFINED STRUCTURE OF THE CYTOCHROME B REDUCTASE FRAGMENT AT 2.5 ANGSTROMS, ITS ADP COMPLEX AND AN ACTIVE SITE MUTANT AND MODELING OF THE CYTOCHROME B DOMAIN |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)