













| General Information: |
|
| Name(s) found: |
IFRH_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 310 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA]
|
| Biological Process: |
response to oxidative stress
[IEP][IGI]
response to cadmium ion [IEP] |
| Molecular Function: |
catalytic activity
[IEA]
transcription repressor activity [IEA] binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATEKSKILV IGGTGYIGKF LVEASAKAGH STFALVREAT LSDPVKGKTV QSFKDLGVTI 60
61 LHGDLNDHES LVKAIKQVDV VISTVGSMQI LDQTKIISAI KEAGNVKRFL PSEFGVDVDR 120
121 TSAVEPAKSA FAGKIQIRRT IEAEGIPYTY AVTGCFGGYY LPTLVQFEPG LTSPPRDKVT 180
181 ILGDGNAKAV INKEEDIAAY TIKAVDDPRT LNKILYIKPS NNTLSMNEIV TLWEKKIGKS 240
241 LEKTHLPEEQ LLKSIQESPI PINVVLSINH AVFVNGDTNI SIEPSFGVEA SELYPDVKYT 300
301 SVDEYLSYFA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
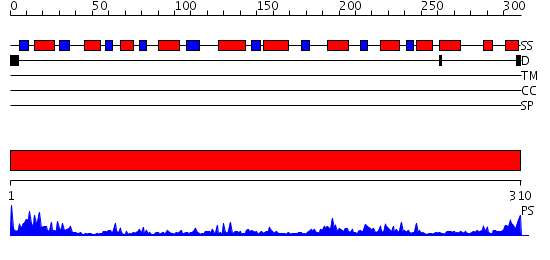
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..310] | 47.221849 | Crystal structures of pinoresinol-lariciresinol and phenylcoumaran benzylic ether reductases, and their relationship to isoflavone reductases |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)