













| General Information: |
|
| Name(s) found: |
GLGL4_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 523 amino acids |
Gene Ontology: |
|
| Cellular Component: |
glucose-1-phosphate adenylyltransferase complex
[IDA]
|
| Biological Process: |
starch biosynthetic process
[TAS]
|
| Molecular Function: |
glucose-1-phosphate adenylyltransferase activity
[IDA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSSYSFALG TSSSILPKLS FRNVENRFYG EKNNNNGLCK RFGSDLGSKK FRNQKFKHGV 60
61 VYAVATSDNP KKAMTVKTSM FERRKVDPQN VAAIILGGGN GAKLFPLTMR AATPAVPVGG 120
121 CYRLIDIPMS NCINSCINKI FVLTQFNSAS LNRHLARTYF GNGINFGGGF VEVLAATQTP 180
181 GEAGKKWFQG TADAVRKFLW VFEDAKNRNI ENILILSGDH LYRMNYMDFV QSHVDSNADI 240
241 TLSCAPVSES RASNFGLVKI DRGGRVIHFS EKPTGVDLKS MQTDTTMLGL SHQEATDSPY 300
301 IASMGVYCFK TEALLNLLTR QYPSSNDFGS EVIPAAIRDH DVQGYIFRDY WEDIGTIKTF 360
361 YEANLALVEE RPKFEFYDPE TPFYTSPRFL PPTKAEKCRM VDSIISHGCF LRECSVQRSI 420
421 IGERSRLDYG VELQDTLMLG ADYYQTESEI ASLLAEGKVP IGIGKDTKIR KCIIDKNAKI 480
481 GKNVIIMNKG DVQEADRPEE GFYIRSGITV IVEKATIQDG TVI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
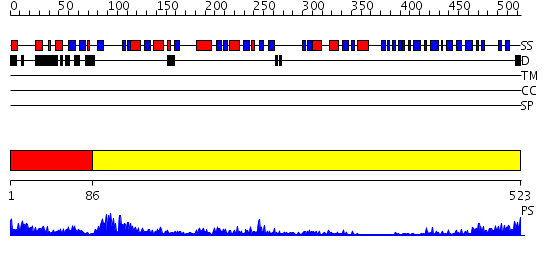
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..85] | 5.045757 | Crystal structure of Inosine-5'-monophosphate dehydrogenase (TM1347) from THERMOTOGA MARITIMA at 2.18 A resolution | |
| 2 | View Details | [86..523] | 68.09691 | Crystal structure of potato tuber ADP-glucose pyrophosphorylase |
Functions predicted (by domain):
| # | Gene Ontology predictions |
| 1 | No functions predicted. |
| 2 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)