













| General Information: |
|
| Name(s) found: |
GIGAN_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1173 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
nucleoplasm [IDA] |
| Biological Process: |
response to cold
[IMP]
flower development [TAS] response to far red light [IMP] positive regulation of long-day photoperiodism, flowering [IMP] response to hydrogen peroxide [IMP] regulation of circadian rhythm [IMP] circadian rhythm [IDA] response to blue light [IDA] regulation of transcription [IDA] temperature compensation of the circadian clock [IMP] |
| Molecular Function: |
protein binding
[IPI]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASSSSSERW IDGLQFSSLL WPPPRDPQQH KDQVVAYVEY FGQFTSEQFP DDIAELVRHQ 60
61 YPSTEKRLLD DVLAMFVLHH PEHGHAVILP IISCLIDGSL VYSKEAHPFA SFISLVCPSS 120
121 ENDYSEQWAL ACGEILRILT HYNRPIYKTE QQNGDTERNC LSKATTSGSP TSEPKAGSPT 180
181 QHERKPLRPL SPWISDILLA APLGIRSDYF RWCSGVMGKY AAGELKPPTI ASRGSGKHPQ 240
241 LMPSTPRWAV ANGAGVILSV CDDEVARYET ATLTAVAVPA LLLPPPTTSL DEHLVAGLPA 300
301 LEPYARLFHR YYAIATPSAT QRLLLGLLEA PPSWAPDALD AAVQLVELLR AAEDYASGVR 360
361 LPRNWMHLHF LRAIGIAMSM RAGVAADAAA ALLFRILSQP ALLFPPLSQV EGVEIQHAPI 420
421 GGYSSNYRKQ IEVPAAEATI EATAQGIASM LCAHGPEVEW RICTIWEAAY GLIPLNSSAV 480
481 DLPEIIVATP LQPPILSWNL YIPLLKVLEY LPRGSPSEAC LMKIFVATVE TILSRTFPPE 540
541 SSRELTRKAR SSFTTRSATK NLAMSELRAM VHALFLESCA GVELASRLLF VVLTVCVSHE 600
601 AQSSGSKRPR SEYASTTENI EANQPVSNNQ TANRKSRNVK GQGPVAAFDS YVLAAVCALA 660
661 CEVQLYPMIS GGGNFSNSAV AGTITKPVKI NGSSKEYGAG IDSAISHTRR ILAILEALFS 720
721 LKPSSVGTPW SYSSSEIVAA AMVAAHISEL FRRSKALTHA LSGLMRCKWD KEIHKRASSL 780
781 YNLIDVHSKV VASIVDKAEP LEAYLKNTPV QKDSVTCLNW KQENTCASTT CFDTAVTSAS 840
841 RTEMNPRGNH KYARHSDEGS GRPSEKGIKD FLLDASDLAN FLTADRLAGF YCGTQKLLRS 900
901 VLAEKPELSF SVVSLLWHKL IAAPEIQPTA ESTSAQQGWR QVVDALCNVV SATPAKAAAA 960
961 VVLQAERELQ PWIAKDDEEG QKMWKINQRI VKVLVELMRN HDRPESLVIL ASASDLLLRA1020
1021 TDGMLVDGEA CTLPQLELLE ATARAIQPVL AWGPSGLAVV DGLSNLLKCR LPATIRCLSH1080
1081 PSAHVRALST SVLRDIMNQS SIPIKVTPKL PTTEKNGMNS PSYRFFNAAS IDWKADIQNC1140
1141 LNWEAHSLLS TTMPTQFLDT AARELGCTIS LSQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
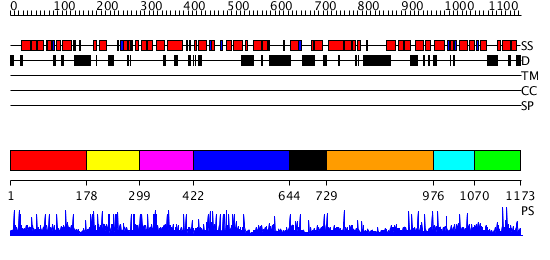
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..177] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [178..298] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [299..421] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [422..643] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [644..728] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [729..975] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [976..1069] | 1.008972 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [1070..1173] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.47 |
Source: Reynolds et al. (2008)