













| General Information: |
|
| Name(s) found: |
FUM1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 492 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA]
|
| Biological Process: |
response to oxidative stress
[IDA]
response to salt stress [IEP] pollen tube development [IMP] |
| Molecular Function: |
protein binding
[IPI]
fumarate hydratase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSIYVASRRL SGGTTVTALR YATSLRSYST SFREERDTFG PIQVPSDKLW GAQTQRSLQN 60
61 FEIGGERERM PEPIVRAFGV LKKCAAKVNM EYGLDPTIGK AIMQAAQEVA EGKLNDHFPL 120
121 VVWQTGSGTQ SNMNANEVIA NRAAEILGRK RGEKCVHPND HVNRSQSSND TFPTVMHIAA 180
181 ATEINSRLIP SLKTLHSTLE SKSFEFKDIV KIGRTHTQDA TPLTLGQEFG GYATQVKYGL 240
241 NRVTCTLPRL YQLAQGGTAV GTGLNTKKGF DVKIAAAVAE ETNLPFVTAE NKFEALAAHD 300
301 ACVETSGSLN TIATSLMKIA NDIRFLGSGP RCGLGELVLP ENEPGSSIMP GKVNPTQCEA 360
361 LTMVCAQVMG NHVAVTVGGS NGHFELNVFK PVIASALLHS VRLIADASAS FEKNCVRGIE 420
421 ANRERISKLL HESLMLVTSL NPKIGYDNAA AVAKKAHKEG CTLKEAALNL GVLTAEEFDT 480
481 LVVPEKMIGP SD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
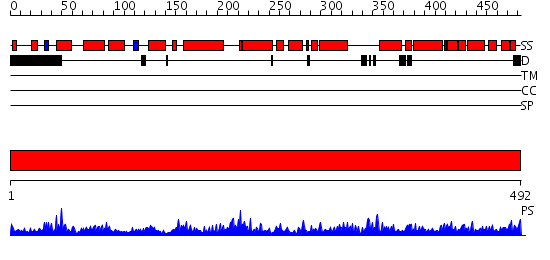
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..492] | 156.0 | RECOMBINANT YEAST FUMARASE |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.74 |
Source: Reynolds et al. (2008)