













| General Information: |
|
| Name(s) found: |
EMF2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 631 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][ISS]
|
| Biological Process: |
regulation of gene expression by genetic imprinting
[IMP]
negative regulation of flower development [IMP] |
| Molecular Function: |
DNA binding
[ISS]
transcription factor activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPGIPLVSRE TSSCSRSTEQ MCHEDSRLRI SEEEEIAAEE SLAAYCKPVE LYNIIQRRAI 60
61 RNPLFLQRCL HYKIEAKHKR RIQMTVFLSG AIDAGVQTQK LFPLYILLAR LVSPKPVAEY 120
121 SAVYRFSRAC ILTGGLGVDG VSQAQANFLL PDMNRLALEA KSGSLAILFI SFAGAQNSQF 180
181 GIDSGKIHSG NIGGHCLWSK IPLQSLYASW QKSPNMDLGQ RVDTVSLVEM QPCFIKLKSM 240
241 SEEKCVSIQV PSNPLTSSSP QQVQVTISAE EVGSTEKSPY SSFSYNDISS SSLLQIIRLR 300
301 TGNVVFNYRY YNNKLQKTEV TEDFSCPFCL VKCASFKGLR YHLPSTHDLL NFEFWVTEEF 360
361 QAVNVSLKTE TMISKVNEDD VDPKQQTFFF SSKKFRRRRQ KSQVRSSRQG PHLGLGCEVL 420
421 DKTDDAHSVR SEKSRIPPGK HYERIGGAES GQRVPPGTSP ADVQSCGDPD YVQSIAGSTM 480
481 LQFAKTRKIS IERSDLRNRS LLQKRQFFHS HRAQPMALEQ VLSDRDSEDE VDDDVADFED 540
541 RRMLDDFVDV TKDEKQMMHM WNSFVRKQRV LADGHIPWAC EAFSRLHGPI MVRTPHLIWC 600
601 WRVFMVKLWN HGLLDARTMN NCNTFLEQLQ I |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
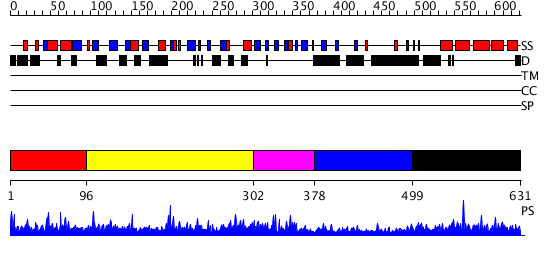
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..95] | 1.010977 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [96..301] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [302..377] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [378..498] | 1.027992 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [499..631] | 4.028986 | View MSA. Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)