













| General Information: |
|
| Name(s) found: |
DNMT1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1534 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
DNA mediated transformation
[IMP]
regulation of gene expression by genetic imprinting [IMP] DNA methylation on cytosine within a CG sequence [IMP] maintenance of DNA methylation [IMP] negative regulation of flower development [IMP] zygote asymmetric cytokinesis in the embryo sac [IMP] |
| Molecular Function: |
methyltransferase activity
[TAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVENGAKAAK RKKRPLPEIQ EVEDVPRTRR PRRAAACTSF KEKSIRVCEK SATIEVKKQQ 60
61 IVEEEFLALR LTALETDVED RPTRRLNDFV LFDSDGVPQP LEMLEIHDIF VSGAILPSDV 120
121 CTDKEKEKGV RCTSFGRVEH WSISGYEDGS PVIWISTELA DYDCRKPAAS YRKVYDYFYE 180
181 KARASVAVYK KLSKSSGGDP DIGLEELLAA VVRSMSSGSK YFSSGAAIID FVISQGDFIY 240
241 NQLAGLDETA KKHESSYVEI PVLVALREKS SKIDKPLQRE RNPSNGVRIK EVSQVAESEA 300
301 LTSDQLVDGT DDDRRYAILL QDEENRKSMQ QPRKNSSSGS ASNMFYIKIN EDEIANDYPL 360
361 PSYYKTSEEE TDELILYDAS YEVQSEHLPH RMLHNWALYN SDLRFISLEL LPMKQCDDID 420
421 VNIFGSGVVT DDNGSWISLN DPDSGSQSHD PDGMCIFLSQ IKEWMIEFGS DDIISISIRT 480
481 DVAWYRLGKP SKLYAPWWKP VLKTARVGIS ILTFLRVESR VARLSFADVT KRLSGLQAND 540
541 KAYISSDPLA VERYLVVHGQ IILQLFAVYP DDNVKRCPFV VGLASKLEDR HHTKWIIKKK 600
601 KISLKELNLN PRAGMAPVAS KRKAMQATTT RLVNRIWGEF YSNYSPEDPL QATAAENGED 660
661 EVEEEGGNGE EEVEEEGENG LTEDTVPEPV EVQKPHTPKK IRGSSGKREI KWDGESLGKT 720
721 SAGEPLYQQA LVGGEMVAVG GAVTLEVDDP DEMPAIYFVE YMFESTDHCK MLHGRFLQRG 780
781 SMTVLGNAAN ERELFLTNEC MTTQLKDIKG VASFEIRSRP WGHQYRKKNI TADKLDWARA 840
841 LERKVKDLPT EYYCKSLYSP ERGGFFSLPL SDIGRSSGFC TSCKIREDEE KRSTIKLNVS 900
901 KTGFFINGIE YSVEDFVYVN PDSIGGLKEG SKTSFKSGRN IGLRAYVVCQ LLEIVPKESR 960
961 KADLGSFDVK VRRFYRPEDV SAEKAYASDI QELYFSQDTV VLPPGALEGK CEVRKKSDMP1020
1021 LSREYPISDH IFFCDLFFDT SKGSLKQLPA NMKPKFSTIK DDTLLRKKKG KGVESEIESE1080
1081 IVKPVEPPKE IRLATLDIFA GCGGLSHGLK KAGVSDAKWA IEYEEPAGQA FKQNHPESTV1140
1141 FVDNCNVILR AIMEKGGDQD DCVSTTEANE LAAKLTEEQK STLPLPGQVD FINGGPPCQG1200
1201 FSGMNRFNQS SWSKVQCEMI LAFLSFADYF RPRYFLLENV RTFVSFNKGQ TFQLTLASLL1260
1261 EMGYQVRFGI LEAGAYGVSQ SRKRAFIWAA APEEVLPEWP EPMHVFGVPK LKISLSQGLH1320
1321 YAAVRSTALG APFRPITVRD TIGDLPSVEN GDSRTNKEYK EVAVSWFQKE IRGNTIALTD1380
1381 HICKAMNELN LIRCKLIPTR PGADWHDLPK RKVTLSDGRV EEMIPFCLPN TAERHNGWKG1440
1441 LYGRLDWQGN FPTSVTDPQP MGKVGMCFHP EQHRILTVRE CARSQGFPDS YEFAGNINHK1500
1501 HRQIGNAVPP PLAFALGRKL KEALHLKKSP QHQP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
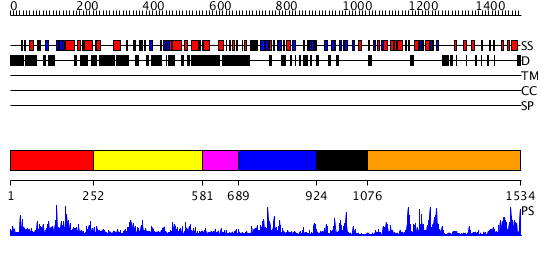
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..251] | 1.084977 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [252..580] | 15.69897 | No description for 2r10A was found. | |
| 3 | View Details | [581..688] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [689..923] | 30.69897 | Crystal structure of the proximal BAH domain of polybromo | |
| 5 | View Details | [924..1075] | 1.153991 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [1076..1534] | 55.221849 | DNA methylase HaeIII |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)