













| General Information: |
|
| Name(s) found: |
PDS_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 566 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA][TAS]
chloroplast envelope [IDA] |
| Biological Process: |
carotenoid biosynthetic process
[IDA]
carotene biosynthetic process [IDA] |
| Molecular Function: |
phytoene dehydrogenase activity
[IDA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVVFGNVSAA NLPYQNGFLE ALSSGGCELM GHSFRVPTSQ ALKTRTRRRS TAGPLQVVCV 60
61 DIPRPELENT VNFLEAASLS ASFRSAPRPA KPLKVVIAGA GLAGLSTAKY LADAGHKPLL 120
121 LEARDVLGGK IAAWKDEDGD WYETGLHIFF GAYPNVQNLF GELGINDRLQ WKEHSMIFAM 180
181 PSKPGEFSRF DFPDVLPAPL NGIWAILRNN EMLTWPEKIK FAIGLLPAMV GGQAYVEAQD 240
241 GLSVKEWMEK QGVPERVTDE VFIAMSKALN FINPDELSMQ CILIALNRFL QEKHGSKMAF 300
301 LDGNPPERLC MPVVDHIRSL GGEVQLNSRI KKIELNDDGT VKSFLLTNGS TVEGDAYVFA 360
361 APVDILKLLL PDPWKEIPYF KKLDKLVGVP VINVHIWFDR KLKNTYDHLL FSRSNLLSVY 420
421 ADMSLTCKEY YDPNRSMLEL VFAPAEEWIS RTDSDIIDAT MKELEKLFPD EISADQSKAK 480
481 ILKYHVVKTP RSVYKTIPNC EPCRPLQRSP IEGFYLAGDY TKQKYLASME GAVLSGKFCS 540
541 QSIVQDYELL AASGPRKLSE ATVSSS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
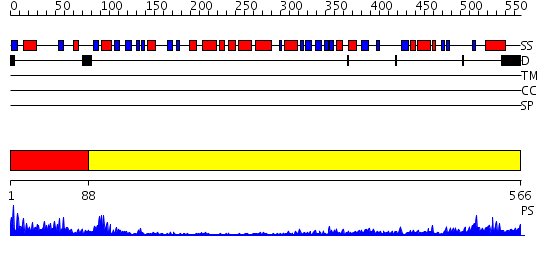
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..87] | 42.0 | The Crystal Structure and Reaction Mechanism of E. coli 2,4-Dienoyl CoA Reductase | |
| 2 | View Details | [88..566] | 59.221849 | The structure basis of specific recognitions for substrates and inhibitors of rat monoamine oxidase A |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)