













| General Information: |
|
| Name(s) found: |
ARFC_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 608 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
vegetative phase change
[IGI][IMP]
regulation of transcription, DNA-dependent [TAS] auxin metabolic process [TAS] abaxial cell fate specification [IGI] response to auxin stimulus [ISS] |
| Molecular Function: |
DNA binding
[ISS]
transcription factor activity [ISS][TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGGLIDLNVM ETEEDETQTQ TPSSASGSVS PTSSSSASVS VVSSNSAGGG VCLELWHACA 60
61 GPLISLPKRG SLVLYFPQGH LEQAPDFSAA IYGLPPHVFC RILDVKLHAE TTTDEVYAQV 120
121 SLLPESEDIE RKVREGIIDV DGGEEDYEVL KRSNTPHMFC KTLTASDTST HGGFSVPRRA 180
181 AEDCFPPLDY SQPRPSQELL ARDLHGLEWR FRHIYRGQPR RHLLTTGWSA FVNKKKLVSG 240
241 DAVLFLRGDD GKLRLGVRRA SQIEGTAALS AQYNQNMNHN NFSEVAHAIS THSVFSISYN 300
301 PKASWSNFII PAPKFLKVVD YPFCIGMRFK ARVESEDASE RRSPGIISGI SDLDPIRWPG 360
361 SKWRCLLVRW DDIVANGHQQ RVSPWEIEPS GSISNSGSFV TTGPKRSRIG FSSGKPDIPV 420
421 SEGIRATDFE ESLRFQRVLQ GQEIFPGFIN TCSDGGAGAR RGRFKGTEFG DSYGFHKVLQ 480
481 GQETVPAYSI TDHRQQHGLS QRNIWCGPFQ NFSTRILPPS VSSSPSSVLL TNSNSPNGRL 540
541 EDHHGGSGRC RLFGFPLTDE TTAVASATAV PCVEGNSMKG ASAVQSNHHH SQGRDIYAMR 600
601 DMLLDIAL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
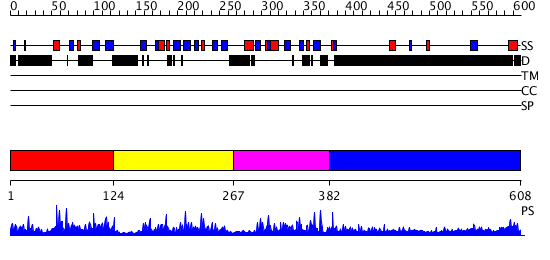
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..123] | 1.081975 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [124..266] | 32.154902 | Solution Structure of the B3 DNA-Binding Domain of RAV1 | |
| 3 | View Details | [267..381] | 3.099989 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [382..608] | 5.089997 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)