













| General Information: |
|
| Name(s) found: |
BAM5_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 498 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endomembrane system
[IEA]
|
| Biological Process: |
response to herbivore
[IEP]
starch catabolic process [IDA] |
| Molecular Function: |
beta-amylase activity
[IDA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATNYNEKLL LNYVPVYVML PLGVVNVENV FADPETLETQ LKRLKEEAGV DGVMVDVWWG 60
61 IIESKGPKQY DWTAYKTLFQ LIARLGLKIQ AIMSFHQCGG NVGDIVTIPI PQWVRDVGDN 120
121 DPDIYYTNRK GTRDIEYLSI GVDNLPLFAG RTAVQLYSDY MSSFKENMAD LIEAGVIVDI 180
181 EVGLGPAGEL RYPSYPQSQG WVFPGIGEFQ CYDKYLKKDF KEAAAKAGHP EWDLPEDAGE 240
241 YNDKPEETGF FKKDGTYVSE KGKFFMTWYS NKLIFHGDQI LGEANKIFAG LKVNLAAKVS 300
301 GIHWLYNHHS HAAELTAGYY NLFKRDGYRP IARMLSKHYG ILNFTCLEMK DTDNTAEALS 360
361 APQELVQEVL SKAWKEGIEV AGENALETYG AKGYNQILLN ARPNGVNPNG KPKLRMYGFT 420
421 YLRLSDTVFQ ENNFELFKKL VRKMHADQDY CGDAAKYGHE IVPLKTSNSQ LTLEDIADAA 480
481 QPSGAFKWDS ETDLKVDG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
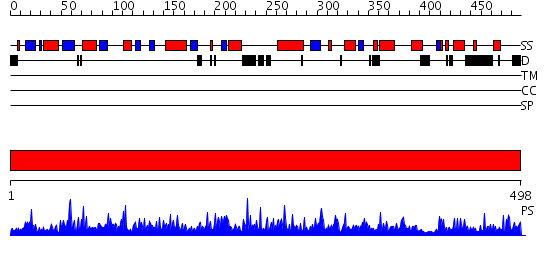
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..498] | 1000.0 | Crystal structure of soybean beta-amylase mutant substituted at surface region |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)