













| General Information: |
|
| Name(s) found: |
HMA5_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 995 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IEA]
integral to membrane [IEA] |
| Biological Process: |
detoxification of copper ion
[IMP]
response to copper ion [IMP] |
| Molecular Function: |
ATPase activity, coupled to transmembrane movement of ions, phosphorylative mechanism
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATKLLSLTC IRKERFSERY PLVRKHLTRS RDGGGGSSSE TAAFEIDDPI SRAVFQVLGM 60
61 TCSACAGSVE KAIKRLPGIH DAVIDALNNR AQILFYPNSV DVETIRETIE DAGFEASLIE 120
121 NEANERSRQV CRIRINGMTC TSCSSTIERV LQSVNGVQRA HVALAIEEAE IHYDPRLSSY 180
181 DRLLEEIENA GFEAVLISTG EDVSKIDLKI DGELTDESMK VIERSLEALP GVQSVEISHG 240
241 TDKISVLYKP DVTGPRNFIQ VIESTVFGHS GHIKATIFSE GGVGRESQKQ GEIKQYYKSF 300
301 LWSLVFTVPV FLTAMVFMYI PGIKDLLMFK VINMLTVGEI IRCVLATPVQ FVIGWRFYTG 360
361 SYKALRRGSA NMDVLIALGT NAAYFYSLYT VLRAATSPDF KGVDFFETSA MLISFIILGK 420
421 YLEVMAKGKT SQAIAKLMNL APDTAILLSL DKEGNVTGEE EIDGRLIQKN DVIKIVPGAK 480
481 VASDGYVIWG QSHVNESMIT GEARPVAKRK GDTVIGGTLN ENGVLHVKVT RVGSESALAQ 540
541 IVRLVESAQL AKAPVQKLAD RISKFFVPLV IFLSFSTWLA WFLAGKLHWY PESWIPSSMD 600
601 SFELALQFGI SVMVIACPCA LGLATPTAVM VGTGVGASQG VLIKGGQALE RAHKVNCIVF 660
661 DKTGTLTMGK PVVVKTKLLK NMVLREFYEL VAATEVNSEH PLAKAIVEYA KKFRDDEENP 720
721 AWPEACDFVS ITGKGVKATV KGREIMVGNK NLMNDHKVII PDDAEELLAD SEDMAQTGIL 780
781 VSINSELIGV LSVSDPLKPS AREAISILKS MNIKSIMVTG DNWGTANSIA REVGIDSVIA 840
841 EAKPEQKAEK VKELQAAGHV VAMVGDGIND SPALVAADVG MAIGAGTDIA IEAADIVLMK 900
901 SNLEDVITAI DLSRKTFSRI RLNYVWALGY NLMGIPIAAG VLFPGTRFRL PPWIAGAAMA 960
961 ASSVSVVCCS LLLKNYKRPK KLDHLEIREI QVERV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
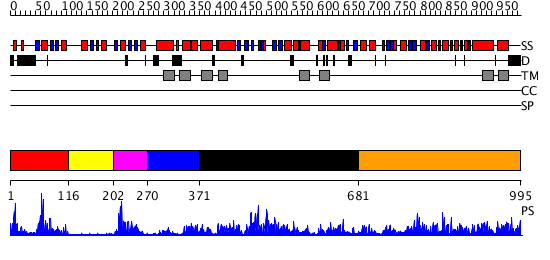
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..115] | 26.0 | No description for 2rmlA was found. | |
| 2 | View Details | [116..201] | 8.853872 | No description for PF00403.18 was found. No confident structure predictions are available. | |
| 3 | View Details | [202..269] | 11.0 | Solution structure of the apo form of the sixth soluble domain A69P mutant of Menkes protein | |
| 4 | View Details | [270..370] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [371..680] | 57.69897 | Calcium ATPase, transduction domain A; Calcium ATPase, catalytic domain P; Calcium ATPase; Calcium ATPase, transmembrane domain M | |
| 6 | View Details | [681..995] | 59.69897 | CopA ATP Binding Domain |
| Source: Reynolds et al. 2008. Manuscript submitted |

|