













| General Information: |
|
| Name(s) found: |
CE34758
[WormBase]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 763 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IEA |
| Biological Process: | NONE FOUND |
| Molecular Function: |
GTP binding
[IEA hydrolase activity, acting on acid anhydrides [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGHKTSKNEK VENHVAFPTA KTLADLPNIT ITHPTPIYQP DTPNPFTVPP PTPRDTHSLR 60
61 SFIEETPILL GSSEHSGSSS PPRSSPPPPP SIPPPSDPPK DGIFHHEDTR KIFGSSVLTR 120
121 GASPLPPKPD ESHYAPPVIK RPIYYAKDKI QLFEALNTRD RDPPVVPKEK KVEKEELAYA 180
181 VGIPHVKFDF AKQEKKAPNL QDNASVSSIK TTDDYPIYNV LNDYNYSSGK TFHSGAFPID 240
241 ANRKNFGFCG RSGSGKSSLI NSLRGLNNGD PQSAGRSHCD RMEPFRFIEG EFQQIVLWEI 300
301 PYPRTFSSSS VVFDANMGFE KLYESHKLKL FKRLFILIPD GAPTDEDITF ARVALSRRTS 360
361 ITFLLTKSDE DLDAENRENG TKLDQAMKRS YETSARLVFS RYLLSKAQIL NDVELLFVNA 420
421 PTARNLVSGT VGYLHYLMNE ERLLELLDLN TGCHYELEVR LRKERDNTET TKPINGNIET 480
481 YRLEEKQIPQ AQAYAVSRPT ILADAGFEIS FGTDDRIYQS LEPKTMIRRA GKTCFNYGFI 540
541 GGRGVGKSSL IDAMRGMSSK NPLSATKLNN RSKAGSCERF EFDDNVLKYS VTLYELSYPK 600
601 KISSYFEFID LVNVASFTAL FILVDQTPSE QDLAFAKIAY RRNTTILFLI SKCDKKLAAR 660
661 SRSDEIPVCD LLKQRYIDKA LQKFDNIMAD KAAELRGRIN VFFVSAPVFK ALRMGDPRES 720
721 QFVLHERAMF DFLKSRRMIA DMLDDPPGEG VYAQLDLDTA GVH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
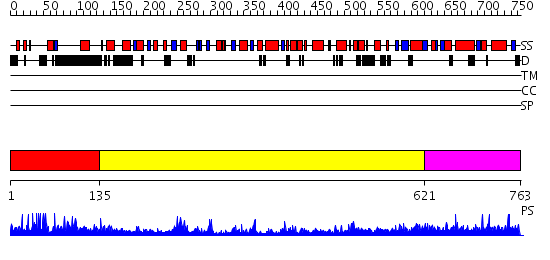
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..134] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [135..620] | 73.69897 | Crystal structure of IIGP1: a paradigm for interferon inducible p47 resistance GTPases | |
| 3 | View Details | [621..763] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
|||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)