













| General Information: |
|
| Name(s) found: |
zfp-1 /
CE33653
[WormBase]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 635 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chromatin
[ISS |
| Biological Process: |
hermaphrodite genitalia development
[IMP embryonic development ending in birth or egg hatching [IMP RNA interference [IMP |
| Molecular Function: |
zinc ion binding
[IEA protein binding [IEA chromatin binding [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLADPLPIKS NKVNNVLGLG SAINAVSLEE RPASGSTVNS GIFVPPPTAF SPPLTTSSRS 60
61 SVAQDPSPPL TINKNSLSSS GPLIPSTAHL SATTASATPI MANGSTLPPS SETTVGTHCL 120
121 QQLQIQSAAA AAITQNQIGP SELNGYPAAS QLSSFMHEIP ARNTTSVASL LPPGAAEYHL 180
181 NGSGDEEKTV KAVLTAPLTK AKRIRDSKND MMDKTHKRPR ANARPPAVLG SMSSGSSGGT 240
241 VGKSPSMQRL QNLVAPIVSE TVTDFQRDRV ADRTAAERRA AAAQSQPSTS TNGGPNVTIP 300
301 AVVEVHTNST NSTNHQNNGL TQNAPASTSM QAGTSSNDGV ISQNGTSSTS QSNRLNLPSF 360
361 MEQLLERQWD QGSSLLMANA HFDVAQLLSC LFQLKSENFR LEENLSGLRK RRDHLFALNS 420
421 RLAEVNTLDV SKRQRSDGLL LQQQIAHHLD PTSIVPKAEV HKQEPLSAPT SVPLPANHSS 480
481 SLFEDIKAPK ATYSRKTPSN IPLTVPLSTA ATALTTTTAA SSGAPVNSNI QNHRATPSTA 540
541 GAPMAATPIM TAVTSANELA ALSPERAQAL LNMYRMPLDA NVAAQLSMIT NFPGQVNPSL 600
601 FSRLLAVNMM NGGLQPNGQP LSALPPPTSA TPNGK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
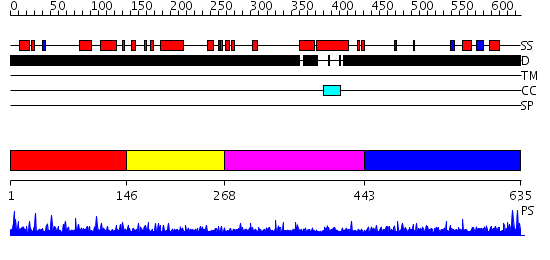
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..145] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [146..267] | 5.16399 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [268..442] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [443..635] | 4.153996 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)