













| General Information: |
|
| Name(s) found: |
CE33035
[WormBase]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 764 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
locomotory behavior
[IMP nematode larval development [IMP positive regulation of growth rate [IMP purine ribonucleoside monophosphate biosynthetic process [IEA growth [IMP |
| Molecular Function: |
AMP deaminase activity
[IEA adenosine deaminase activity [IEA deaminase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNNLPPDITE AQIETVDSEL FDSGPFKKNF AFGNADHSTA SPFEISTTPI EVNESRAHKT 60
61 IEISNREKGA KSRRWSCLAR LESQNDGCLS PSSLKSVLDE IQTEPRCKSP QHIDLHTFRD 120
121 AVDVNYQRMA ITGEELSGVP LEDLKTASGH LIEALHLRSK YMERIGNQFP STTRNFLSGH 180
181 YPANLPKHRV KNTETTVQTS FNPPDPPKDH WGKNDPLPKY EKIYHLRRNR GVTEICNDDG 240
241 SIDQQFKNVN VTKEEFLNDT EKLTAMIVDG PLKSFCFRRL SYLENKFQLH VLLNELRELH 300
301 EQKGVSHRDF YNIRKVDTHI HAASSMNQKH LLRFIKKKIK TEADTVVLNN NGTKVTMKEV 360
361 FKKMGIDAYD LSVDMLDVHA DRNTFHRFDK FNTKYNPVGE STLREIFIKT DNYVGGKYFA 420
421 DLLKEVLSDL EDSKYQHAEP RLSIYGRSKN EWDNLAKWAL THDVWSPNAR WLVQIPRLYD 480
481 VYRAKNMVKN FDDMLDNLFT PLFEVTNDPS SHPELHLFLQ QISGIDSVDD ESKHEFVNFD 540
541 RSTPCPPEYT DLENPPYNYY LFYMYRNICA LNAFRRARGL NTFALRPHCG EAGHVSHLLT 600
601 GYLTSESIAH GILLRKVPVL QYLYYLTQIG IAMSPLSNNS LFISYQRNPL PEYLQKGLNV 660
661 SLSTDDPLQF HYTKEALMEE FSIAAQVWKL SSCDMCELAR NSVMQSGFED KVKIHWLGPN 720
721 YKEEGVLGND IHRTNVPDIR VSFRHEALVD ELYNLFRVQN TLKQ |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
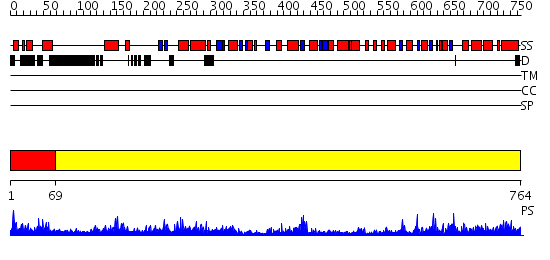
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..68] | 1.042995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [69..764] | 163.0 | X-Ray Structure of Adenosine 5'-Monophosphate Deaminase from Arabidopsis Thaliana in Complex with Coformycin 5'-Phosphate |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)