













| General Information: |
|
| Name(s) found: |
CE32364
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 400 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
negative regulation of multicellular organism growth
[IMP nematode larval development [IMP positive regulation of growth rate [IMP embryonic development ending in birth or egg hatching [IMP biosynthetic process [IEA growth [IMP |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVRVTILHPD LGIGGAERLI VDAAVGLQDR GHSVRIFTNQ YSRSHCFQET LDLDICTVVP 60
61 WIPRSIFGKG HALCAYLKMI IAALYIVIYH KDTDVILSDS VSASQFVLRH FSKAKLVFYC 120
121 HFPDRLLTKR DGNLKAFYRN IIDWIEEYTT GLADVICVNS NFTKNVVRET FKSLASQELT 180
181 VLYPSLNTEF FDSIEASDDF GEEIPRGTKY VFTSLNRFER KKNIVLALDA FEKLKSNLPA 240
241 DEFSQCHLVI AGGYDLKNPE NIEHYDELVE HMKKLELPAD QIVFLHSPSD TQKVNLIRRS 300
301 RAVLYTPDRE HFGIVPVEAM YLGTPVIAVN TGGPCESVRN NETGFLVDQT AEAFAEKMID 360
361 LMKDEEMYRR MSEEGPKWVQ KVFAFEAFAR KLDEIIQSTL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
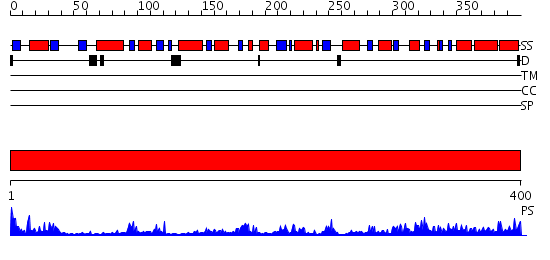
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..400] | 49.154902 | Structure of glycogen synthase from Pyrococcus abyssi |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.82 |
Source: Reynolds et al. (2008)