













| General Information: |
|
| Name(s) found: |
mmcm-1 /
CE30404
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 744 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
metabolic process
[IEA lactate fermentation to propionate and acetate [IEA |
| Molecular Function: |
intramolecular transferase activity
[IEA metal ion binding [IEA cobalamin binding [IEA methylmalonyl-CoA mutase activity [IEA isomerase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYLQLLKPTL LRCSTRPSSS GAYTRSPIDQ KWAAMAKKAM KGREADTLTW NTPEGIPIKP 60
61 LYLRSDRDCD AQRSVELPGQ FPFTRGPYPT MYTQRPWTIR QYAGFSTVEE SNKFYKENIK 120
121 AGQQGLSVAF DLATHRGYDS DNPRVFGDVG MAGVAVDSVE DMRQLFDGIN LEKMSVSMTM 180
181 NGAVVPVLAM YVVAAEEAGV SRKLLAGTIQ NDILKEFMVR NTYIYPPEPS MRIIGDIFAY 240
241 TSREMPKFNS ISISGYHMQE AGADAVLEMA FTIADGIQYC ETGLNAGLTI DAFAPRLSFF 300
301 WGISMNFYME IAKMRAARRL WANLIKERFS PKSDKSMMLR THSQTSGWSL TEQDPYNNII 360
361 RTTIEAMASV FGGTQSLHTN SFDEALGLPT KFSARIARNT QIIIQEESGI CNVADPWGGS 420
421 YMMESLTDEI YEKALAVIKE IDELGGMAKA VASGMTKLKI EEAAAKKQAR IDAGKDVIVG 480
481 VNKYRLDHEQ QVEVLKIDNA KVREEQCAKL NHIRATRDAE KAQKALDAIT EGARGNGNLM 540
541 ELAIEAARAR CTVGEISDAM EKVFNRHAAV NRLVSGAYKS EFGETSEMSQ VLERVKSFAD 600
601 RDGRQPRIMV AKMGQDGHDR GAKVIATGFA DLGFDVDVGP LFQTPLEAAQ QAVDADVHVI 660
661 GASSLAAGHL TLIPQLIGEL KKLGRPDILV VAGGVIPPQD YKELYDAGVA LVFGPGTRLP 720
721 ACANQILEKL EANLPEAPGK AASR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
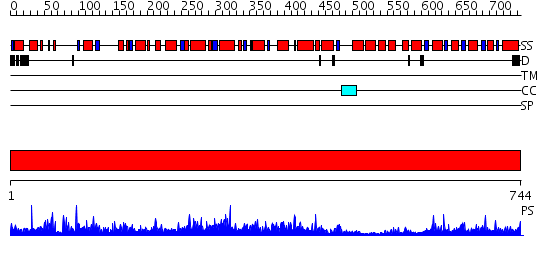
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..744] | 1000.0 | Methylmalonyl-CoA mutase alpha subunit, domain 1; Methylmalonyl-CoA mutase alpha subunit, C-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.76 |
Source: Reynolds et al. (2008)