













| General Information: |
|
| Name(s) found: |
CE30560
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 709 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
metabolic process
[IEA cysteine metabolic process [IEA |
| Molecular Function: |
cysteine desulfurase activity
[IEA pyridoxal phosphate binding [IEA catalytic activity [IEA transaminase activity [IEA molybdenum ion binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPYLDHAGST LPSKIQLEEV AKQQSQLILA NPHSHHATAV KTKQIVNSAR LRILQYFNTT 60
61 SDDYFVVLTN NTTHGLKIVA ENFKFGQKTH SILNIASVLH GGSSNLGYLY DSHHSVVGLR 120
121 HVVNGKVNSI SCVNEESILE HEIPDVEHSL FVLTAMSNFC GKKYSLESVH RLQEKGWAVC 180
181 LDAASFVSSS ALDLSQQRPN FIAFSFYKIF GYPTGIGALL VRKDSAHLIE KTSFAGGTVQ 240
241 SVDEMSMFFV LREFERAFEE GTLNYYGIAQ LQKGFEEIER CGGISSIRNL THHLCKNALY 300
301 MLKSKKHPNG RPVVEIYSQS EQFENPDKQG PIVAFNLKRP DGGYYGYTEV EKMCAIFGIE 360
361 LRTGCFCNIG ACKKYLGITS EMIQENMSKG KRCGDEIDLI NGTPTGAIRI SFGRTSTEHD 420
421 ITALEQMIDT CFTEGEHQAQ SKPDPMNIES YSPTVVNLFS FPIKSVGSVG RKRYELTARG 480
481 FKNDREFLIV NDDVTLNLKT HPELCMLTAT IVDDDQLLIQ TFDQNENLVL PMSLSLKDNG 540
541 AKLVCKNTIA TMDCGDKVGK WLDNALDRQN CRLLRVAEDS KKNFVNDSPF LLINEASVYM 600
601 LSRYINMEVR EILTRFRSNI VVRGLPPFIE DTAKRLSIEN LEFEVVDKCT RCEMICVDPM 660
661 TGEKDPSLLL ALRDYRNKQK MTFGIYIRQT NFESGQYLES GMSVNFSTD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
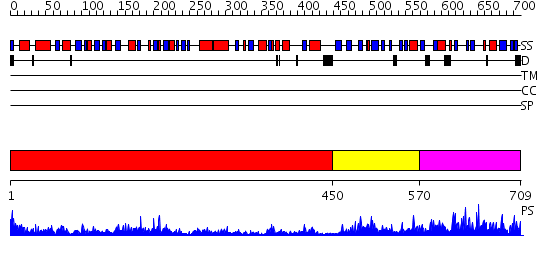
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..449] | 86.69897 | Cysteine desulfurase IscS | |
| 2 | View Details | [450..569] | 12.30103 | Solution structure for the protein coded by gene locus BB0938 of Bordetella bronchiseptica. Northeast Structural Genomics target BoR11. | |
| 3 | View Details | [570..709] | 4.1 | Crystal Structure of APC1665, YUAD protein from Bacillus subtilis |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||
| 1 |
|
||||||||||||
| 2 |
|
||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)