













| General Information: |
|
| Name(s) found: |
ipla-1 /
CE29816
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 753 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: |
metal ion binding
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAQTKDADLS RPSGLPSNGN PGSSTTVPPA SKSGPVAPPR PSGTPVAPQR NRRRKVTELK 60
61 CSEVRWFFQE PKGTLWNPFN GRDSIMLEIK YRKEKGIELD EAMQEIYDES LTHYKMEMKD 120
121 EPEIENGNIG MEQEKPMVVV MNGQYKVNKD NSKIDPIYWK DDSKEIRRGS WFSPDYQPLE 180
181 MPLSDQIEKN HLQCFRNQMI PEGTTVFSKS ETSNKPVLAE LHVDGYDIRW SSVIDISLHQ 240
241 KGNAILRYLW AKSTPLRRGY EKEADWNDAA AEISHLILVV HGIGQKGYEN LIAQNANQVR 300
301 DGVVSAMEKV YPEEKSRPMF LPVEWRSALK LDNGLTDNIT IPKMSSMRAS LNSTAMDVMY 360
361 YQSPLFRTEI VRGVVSQLNR TYKLFKANNP QFNGHVSVFG HSLGSVICYD VLTQYSPLML 420
421 FDKYVTKSID EYLKRDDTNA SEEARKALEA MKLAREQLRD NLEGGIHKLL VTKEEQLEFK 480
481 VKYLFAVGSP LGVFLTMRGG ESTDLLSKAT NVERVFNIFH PYDPVAYRLE PFFAPEYRHI 540
541 RPIKLFSNTD LRARASYENL PLDVYKHYLK KLKNLNKAKK NKDDKTADAR SGGDDENEDE 600
601 DECDSDEDAR SGCSSPRSMT PPPFETAAAN AAAAAKETKA VKKGWFSFGT SSNPKKTQST 660
661 ASLGSVNATS TENIEFAKEA AEELPLAEKI LGSGVRVPHR IDFQLQPALT EKSYWSVLKS 720
721 HFAYWTNADL ALFLANVLYC KPLKPEEAKP TWA |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) | |
| View Run | hcp1 IP from december2004 | Cheeseman, I.M., et al. (2005) | |
| View Run | hcp2 ip from december | Cheeseman, I.M., et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
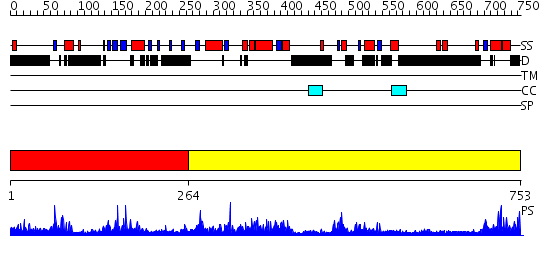
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..263] | 3.084976 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [264..753] | 0.982 | Lipase L1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)