













| General Information: |
|
| Name(s) found: |
cct-6 /
CE30647
[WormBase]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 429 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
hermaphrodite genitalia development
[IMP locomotory behavior [IMP nematode larval development [IMP embryonic development ending in birth or egg hatching [IMP cellular protein metabolic process [IEA protein folding [IEA reproduction [IMP growth [IMP |
| Molecular Function: |
ATP binding
[IEA protein binding [IEA unfolded protein binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSIQCLNPK AELARHAAAL ELNISGARGL QDVMRSNLGP KGTLKMLVSG AGDIKLTKDG 60
61 NVLLHEMAIQ HPTASMIAKA STAQDDVTGD GTTSTVLLIG ELLKQAESLV LEGLHPRIVT 120
121 EGFEWANTKT LELLEKFKKE APVERDLLVE VCRTALRTKL HQKLADHITE CVVDAVLAIR 180
181 RDGEEPDLHM VEKMEMHHDS DMDTTLVRGL VLDHGARHPD MPRHVKDAYI LTCNVSLEYE 240
241 KTEVNSGLFY KTAKEREALL AAEREFITRR VHKIIELKKK VIDNSPDGKN KGFVVINQKG 300
301 IDPPSLDLLA SEGILALRRA KRRNMERLQL AVGGEAVNSV DDLTPEDLGW AGLVYEHSLG 360
361 EEKYTFIEEC RAPKSVTLLI KGPNKHTITQ IKDAIHDGLR AVFNTIVDSC SPWSCCFRNC 420
421 CLRDVEKRC |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
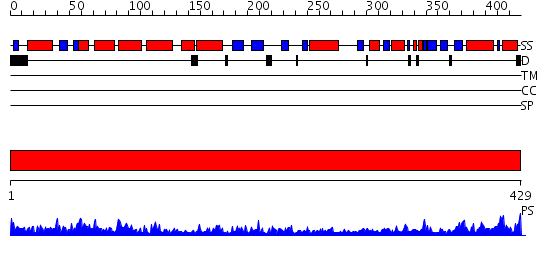
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..429] | 112.0 | Crystal structure of the chaperonin from Thermococcus strain KS-1 (nucleotide-free form of single mutant) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)