













| General Information: |
|
| Name(s) found: |
CE29368
[WormBase]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 725 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
embryonic development ending in birth or egg hatching
[IMP]
meiotic chromosome segregation [IMP] meiosis [IMP] |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPPPSNQSAW SSNLLNARKR RWKKDEEDEN SNTERNASLD LNLSNVELTE ALDRNGDAQI 60
61 TPERRRVSPF VDFDDNLDVE SKRNIDLNLT ITPETDIPFD TQPSTSSIAE SSSSPPRTPH 120
121 SFERSSSPLY SSSDSVGVFS NETPYVKGIR EDLSRERMRF LEVDSILDSI SEKYLQQNDS 180
181 QSFQVQLTHW QTIIFQEKCR FSNNQLRRVK QNYKMFTDLE IMPTINLTMQ RKTRMSTIDN 240
241 YKATTYISKG EKVTVVKIID VEKSVIPKLE RLSVSKQLRH DSYTDGKIVI GIGGDSGGGT 300
301 TKLCLLIGNC DHANSPHRIV LLAVFDDSDS RELIMAYLSD LIVKINNFTS ITYMEDGRVV 360
361 TRGVVQKVVG DFKFTCDLLS HKKQCATYFC PFCFETNPRG GLMQKLKDLN LQKIYFLKTM 420
421 NSYKLNSKYG SFGVRCGSGP ILQNVKLEHY LPAMLHLIVG LFTKYIFEPI WMAVIQDANK 480
481 KFEASPLKRK REMKAQFTAL KEEKVLLDET LDGLAGGYLK QFENDLEEVG ATRRAWFQMY 540
541 TGNHTKLILS EKGVTAAFKN LKNHMTPMLL NVKNAMSRLS KIMSLSANRL LSDDDISELD 600
601 ESMKEFVEFL QAAHPEESIT QKLHVLVAHV VEVAKTERNL GRLSEQGIES LHAVFNRLER 660
661 RFHSVRNTGK RYLTPRLPNK PPSALPHTIL NLFAIKISGF PQYHKIRDFL WLVVKTLTSR 720
721 YSVQI |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
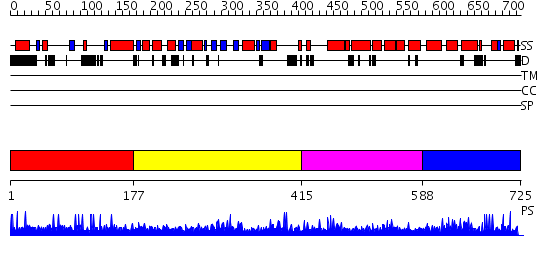
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..176] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [177..414] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [415..587] | 1.01498 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [588..725] | 2.013979 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)