













| General Information: |
|
| Name(s) found: |
wapl-1 /
CE28750
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 748 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
locomotory behavior
[IMP embryonic development ending in birth or egg hatching [IMP positive regulation of growth rate [IMP oviposition [IMP |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSDANSDDP FSKPIVRKRF QATLAQQGIE DDQLPSVRSS DSPDVPDTPD VPVNQLSSPP 60
61 LSLPETLSEG NAETLQNLSD DSEPEMLSQS STSSLNRRME DSAIDPSRGT RKSQSRGFDY 120
121 DPAGERTTAP VQKKKKDEID MGGAKFFPKQ EKKHVYTHKW TTEEDDEDEK TISSSSNRYS 180
181 SRPNQPAVSA RPRQPVYATT STYSKPLASG YGSRVRHIKE ANELRESGEY DDFKQDLVYI 240
241 LSSLQSSDAS MKVKCLSAIS LAKKCVSPDF RQFIKSENMT KSIVKALMDS PEDDLFALAA 300
301 STVLYLLTRD FNSIKIDFPS LRLVSQLLRI EKFEQRPEDK DKVVNMVWEV FNSYIEKQEV 360
361 GGQKVSFDMR KESLTPSSLI IEALVFICSR SVNDDNLKSE LLNLGILQFV VAKIETNVNL 420
421 IADNADDTYS ILILNRCFRI LESSSVFHKK NQAFLISHRS NILISSLAKF LQVILDRVHQ 480
481 LAEEEVKKYI SCLALMCRLL INISHDNELC CSKLGQIEGF LPNAITTFTY LAPKFGKENS 540
541 YDINVMMTSL LTNLVERCNA NRKVLIAQTV KMVIPGHDVE EVPALEAITR LFVYHESQAQ 600
601 IVDADLDREL AFDEGGCGDE EEEEEGGDES SDEDGVRKDG RLDRNKMDRM DQVDVVHALQ 660
661 QVMNKASAHM EGSVIASYHA LLVGFVLQQN EDHLDEVRKH LPGKNFQNMI SQLKRLYDFT 720
721 KATMAKRVES NSGFRAIERV IEYLERLE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
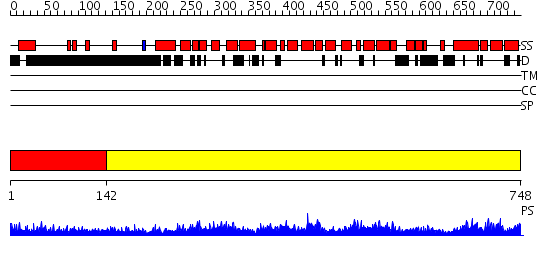
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..141] | 4.246995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [142..748] | 0.985 | Crystal structure of the exportin Cse1p complexed with its cargo ( Kap60p) and RanGTP |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)