













| General Information: |
|
| Name(s) found: |
CE27831
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 484 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
embryonic development ending in birth or egg hatching
[IMP metabolic process [IEA reproduction [IMP |
| Molecular Function: |
nucleotidyltransferase activity
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTITAPPKDE IISKFPGSEP LLNFYNELSD AEKSKLFHQI STLNLSEAHQ WFIDSADQRA 60
61 PSTAEDLKPV LDSQHFVQAE LHQVILDGLW NKGMDAIGRG EVCAIVLAGG QATRLGSSQP 120
121 KGTIPLGINA SFGDSLLGIQ AAKIALLQAL AGEREHQNPG KIHWAVMTSP GTEEATREHV 180
181 KKLAAHHGFD FDEQITIFSQ DEIAAYDEQG NFLLGTKGSV VAAPNGNGGL YSAISAHLPR 240
241 LRAKGIKYFH VYCVDNILCK VADPHFIGFA ISNEADVATK CVPKQKGELV GSVCLDRGLP 300
301 RVVEYSELGA ELAEQKTPDG KYLFGAGSIA NHFFTMDFMD RVCSPSSRLP YHRAHKKISY 360
361 VNEQGTIVKP EKPNGIKLEQ FIFDVFELSK RFFIWEVARN EEFSPLKNAQ SVGTDCLSTC 420
421 QRDLSNVNKL WLERVQAKVT ATEKPIYLKT IVSYNGENLQ ELRHREISDS ALESDHSINK 480
481 FFVV |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
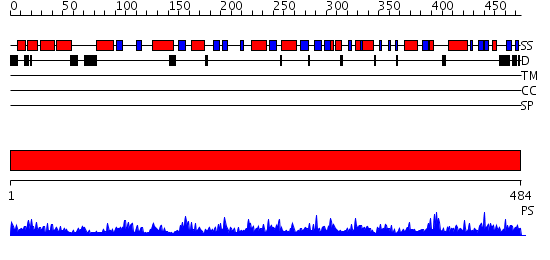
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..484] | 156.0 | UDP-N-acetylglucosamine pyrophosphorylase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)