













| General Information: |
|
| Name(s) found: |
unc-11 /
CE27813
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 548 amino acids |
Gene Ontology: |
|
| Cellular Component: |
presynaptic membrane
[IDA |
| Biological Process: |
locomotion
[IMP synaptic vesicle endocytosis [IMP embryonic development ending in birth or egg hatching [IMP post-embryonic development [IMP gamete generation [IMP |
| Molecular Function: |
clathrin binding
[ISS phospholipid binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQTIEKALHQ PMPFTTGGQT ISDRLTAAKH SLAGSQLGKT ICKATTEEVM APKKKHLDYL 60
61 LHCTNEPNVS IPSMANLLIE RTQNPNWTVV YKALITIHNI MCYGNERFSQ YLASCNTTFN 120
121 LTAFVDKVGG AGGYDMSTHV RRYAKYIGEK INTYRMCAFD FCKVKRGRED GLLRTMHTDK 180
181 LLKTIPILQN QIDALLEFSV TTSELNNGVI NCSFILLFRD LIRLFACYND GIINVLEKYF 240
241 DMNKKQCRDA LDTYKSFLTR LDKVAEFLRV AESVGIDRGE IPDLTRAPAS LLEALEAHLI 300
301 HLEGGKAPPP TQQHVAPHQF TTGFAFSQQP QPALGDAERQ RYIELEQERL RQFEDQKKSI 360
361 NSANPFANDV ASAAPAPATS AAQPDLLDMF QSSAAPAPQT ADVTNPFGNF AAPSAFPTNG 420
421 HQAAPFGYPN AHPDDLARMT AQMSLNQQGY RAPAGWNTTT SAVSNNPFGA TSAPQPMYTA 480
481 PMGMYQQPFG AQPMWNPAMA AYGQQYGYGQ PVPPQQQHQI QLVHAAMAAK NAAQAQQAQA 540
541 ASADPFGL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
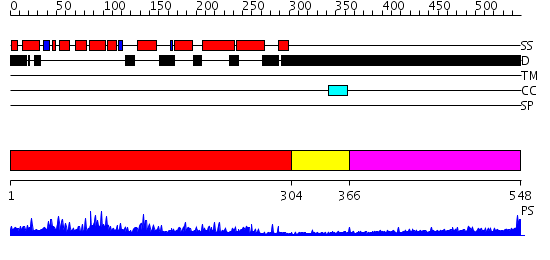
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..303] | 107.0 | Clathrin assembly lymphoid myeloid leukaemia protein, Calm | |
| 2 | View Details | [304..365] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [366..548] | 8.0 | Sec24 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)