













| General Information: |
|
| Name(s) found: |
CE25500
[WormBase]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 710 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IEA |
| Biological Process: |
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
[IEA |
| Molecular Function: |
nucleic acid binding
[IEA 3'-5' exonuclease activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDFMSNTAA LQLLHNAVKS YEREVANFFA ANEKATSKYT GRFDFRQFAK KMFDPIPTAQ 60
61 LGQLYIEMLE ESHIYMHSHQ KSRVGANGYP HSFVSVPELL LNMILAYQLP QQRFVPAEKM 120
121 EEDGGKVEED GGAGGAGGSR KKRRNSAARK GKNVTINPRV VLGNELAAKL LEEMLKIPLA 180
181 PESKQWKTAK YVLCPSSCPQ QMEVFGEIIA KNTKSGLISF NALNYFMHDP ILNRYLDDPI 240
241 TILHALYIGP LQAPTVELFA TNRTDVFRQR CREVLRIALH ASKDPMTFTY SVPEFIDKNH 300
301 PDRCSYRDFV ETVTQLIRDL NGEHQGSTAI HTGWACAAIK KFGQKRYVDK TWKDENYYDL 360
361 IWTVLAQRPH LKKFVVDVLE KNFKDLDAAR FWRRMQPYQM NPTVIFNQNV IIVDPLQPTD 420
421 EFLNFPPTVR HIQMVTNEKE IRDLQMLLEY KVSREEKIYV GVDAEWSAYV SPSKATILQM 480
481 SLHDCIYIID LESYQISPQS YHHVLSYLFE TPEIVKIGFQ FGEDLHQLRA AFRNCVALYR 540
541 PNNVVCVGKL IMDLMDEIGK LENCDDFRKE WLPHLTDNET SSAANSSVEE QANETIDTNE 600
601 SMDEGDGGGS GIVVKKEESV KQQFLNKGLS FICEKLLGRP LDKTEQCSVW DRRPLRCLQL 660
661 RYAAMDAYCM LLLYEKCATI FAKLGINVND FVSKQSPIRI SLPLLSEEKL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
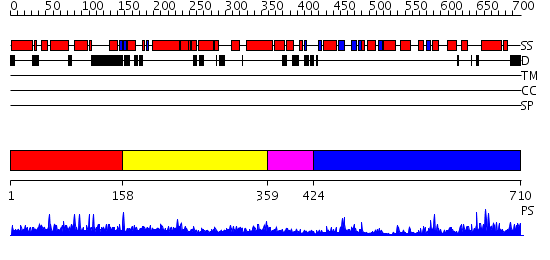
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..157] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [158..358] | 1.030979 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [359..423] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [424..710] | 30.39794 | No description for 2hbjA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)