













| General Information: |
|
| Name(s) found: |
CE25311
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 766 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGDPVEGKGG PQPRGGKKKD ADDEKNAWEE GDFIDEKIHL KLSRGSTCTS SATSSAPSST 60
61 SSTRARRNWS VFSERLRETL SRVGGGGPSS TTDSLILFDV KRVGDCMWIC ERAASPPSSF 120
121 DSSSSDSDDD SFLRIFKFFP PTPAVSLDTL ASGFDNTGNV RIWPGSEALA WTILRDPNKI 180
181 LGPSLGDGDG GGHRRILELG AGFLGLSSIL IACKVPDSSV WVTDGNMESV RSLEKIRNSN 240
241 SSLDRCCHVR QLIWGAQELQ QQHTFDTILA ADCVFFSEHH ESLMKCIHSH LRPSGRAILS 300
301 SPRRKQSLQK FLDYVGTVWS DEFTVCIEPD LNLALEQKIG RIWMDGEEKD EKFPILELKG 360
361 ALQLGVLKTA DSVALRAGER NGLDMVVEED VVSQVGALVW DTVAQDWADD LARFVAHAGR 420
421 QRVNMDDISL LTRRNPDLLR AACELAGVDC PQETSGGTKG KPRKRKKDDV TVAPMLRAAP 480
481 SAPNSQNAPL LAPIQEEDDE NDDDDEITIL DDQILHTPQQ KSYSKKAKFS EEEAPSSRKR 540
541 ISTSTPKTTA ERTLKFPDEI TPIRQPNDLQ EIVVFNRANL SAIEEEEQQD SFDAFGTTGG 600
601 NDSSSKTITN TSSLLRQAEE HRSSIVEPTK DRIEKERLDR LVPDDEEFEN ASVDSFDNFN 660
661 AEPDPAPAPA LAPVPEKRTS VFSKKICSFD FDDDDGDDSF DEPVVVVAEV PKLPVQQQKT 720
721 PKASGSESKK KPEKRPESST PSVFSKKNKT IQLDDHDSFD DFDFGI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
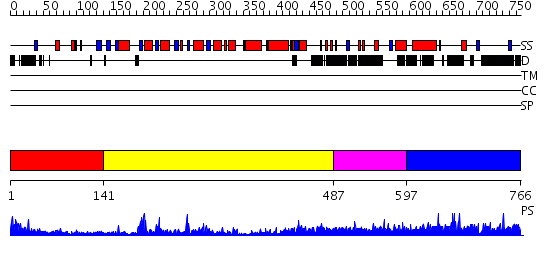
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..140] | 15.30103 | Crystal structure of Tt1595, a putative SAM-dependent methyltransferase from Thermus thermophillus HB8 | |
| 2 | View Details | [141..486] | 50.522879 | Protein arginine N-methyltransferase 1, PRMT1 | |
| 3 | View Details | [487..596] | 1.035991 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [597..766] | 1.029943 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 |
|
|||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)