













| General Information: |
|
| Name(s) found: |
lag-1 /
CE25048
[WormBase]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 671 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
positive regulation of vulval development
[IMP collagen and cuticulin-based cuticle attachment to epithelium [IMP molting cycle, protein-based cuticle [IMP regulation of cell fate specification [IMP hermaphrodite genitalia development [IMP nematode larval development [IMP embryonic development ending in birth or egg hatching [IMP oviposition [IMP germ-line stem cell division [IMP positive regulation of transcription from RNA polymerase II promoter [IMP cell fate specification [IMP |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPLAYSTHDN FYESPKTPQP TWDHVHAQFP LGEPARNLDK FIEAMFQSVS PLAGVAAAPS 60
61 QIAALQQIQA LMTFQMQQNN LFPKIDTISK SPTPELASPS AKRMRLSPST SSHSDVASTS 120
121 KGTNGQDQSK NSPTNSDPFY TIGQSTPNTS SFLDTSNSSF GAPSTVVANP MTNYQLAFQA 180
181 KLGSLHSLIG DSVQSLTSDR MIDFLSNKEK YECVISIFHA KVAQKSYGNE KRFFCPPPCI 240
241 YLIGQGWKLK KDRVAQLYKT LKASAQKDAA IENDPIHEQQ ATELVAYIGI GSDTSERQQL 300
301 DFSTGKVRHP GDQRQDPNIY DYCAAKTLYI SDSDKRKYFD LNAQFFYGCG MEIGGFVSQR 360
361 IKVISKPSKK KQSMKNTDCK YLCIASGTKV ALFNRLRSQT VSTRYLHVEG NAFHASSTKW 420
421 GAFTIHLFDD ERGLQETDNF AVRDGFVYYG SVVKLVDSVT GIALPRLRIR KVDKQQVILD 480
481 ASCSEEPVSQ LHKCAFQMID NELVYLCLSH DKIIQHQATA INEHRHQIND GAAWTIISTD 540
541 KAEYRFFEAM GQVANPISPC PVVGSLEVDG HGEASRVELH GRDFKPNLKV WFGATPVETT 600
601 FRSEESLHCS IPPVSQVRNE QTHWMFTNRT TGDVEVPISL VRDDGVVYSS GLTFSYKSLE 660
661 RHGPCRIVSN Y |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
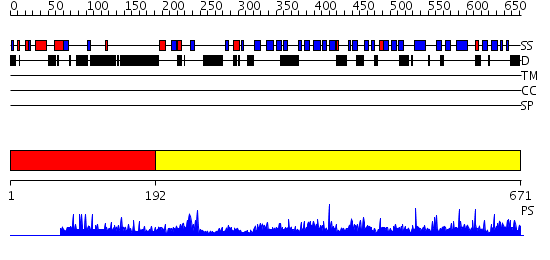
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..191] | 1.022968 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [192..671] | 1000.0 | Crystal Structure of CSL bound to DNA |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.79 |
Source: Reynolds et al. (2008)