













| General Information: |
|
| Name(s) found: |
CE24996
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 742 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IEA |
| Biological Process: |
regulation of small GTPase mediated signal transduction
[IEA signal transduction [IEA |
| Molecular Function: |
GTPase activator activity
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDWIYSWKDS IFYGMCSCSG SPSYHEKDVP HPHDRLDMPL QLKAYRRNSL NAEGYYRDLE 60
61 RMHERGLFTT GINYNAEKSE DFLNMIERMQ SNRLDDQRCE MPERTHNPTS GHQNEQTHQK 120
121 TNGSAVHNQL TRNIMNEVLT KVGPYPQIVL PSNGFWMDGV NQQHHMDDQV NNMNVNSCAR 180
181 FKLETDETSH CYRRHFFGRE HHDFFANDPI VGPLVLSVRT EVISSCDHFR IILRTRKGTI 240
241 HEIVSATALA DRPSASRMAK LLCEEITTEQ FSPVAFPGGS ELIVQYDEHV LTNTYKFGVI 300
301 YQKGGQTTEE QLFGNPHGSP AFDEFLSMIG DSVQLNGFQK YRGGLDTAHN QTGHQSVFSE 360
361 FKNREIMFHV STMLPYTIGD AQQLQRKRHI GNDIVAIIFQ EANTPFAPDM IASNFLHAYV 420
421 VVQPIDALTD RVRYRVSVAA RDDVPFFGPT LPTPSIFKRG QDFRNFLLTK LINAENAAYK 480
481 SSKFAKLAER TRSSLLDGLH ATLRERAEFY ATPLLESTSS GGESAATTSS SSNSYGGGIL 540
541 SSVKKAFIGR SRSVSQEASH VPHRAATINV TSRPKKSISS TSSSTSARTH SPIRDEEVVK 600
601 PYRKEWEISS QESPDNEHDS DTGMESMSST EMSGQTRASS CTFCVDDFHV AANHQSSDAK 660
661 RLETLCVDVT RLQNEKHDLL RQNVSCKTDI KKLKDRQSVL SEELERANDE ISRLRRMLKK 720
721 PSNSDMAPVH QHFERSYSDV SV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
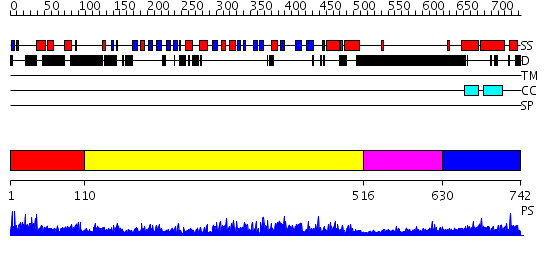
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..109] | 1.046991 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [110..515] | 129.0 | Crystal Structure of the Rap1GAP catalytic domain | |
| 3 | View Details | [516..629] | 3.058995 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [630..742] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)