













| General Information: |
|
| Name(s) found: |
CE23543
[WormBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 746 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
positive regulation of growth rate
[IMP]
|
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRNKRVLKIS DSNSKLRSGG LANIKKSRSM PESPFRTGTP SRMPSAFGEK TYMSRVPRII 60
61 DRNRRATVTT FDLAPIQVSK EEHEKERSRW LGKMEEVEKR LMDAETLNSS LTINKAELKK 120
121 KIVDMEKGQR PMIDANRKLS DRNKVLQQEV KKVEQRFSHS RDDFLTLKDI HERLLKENAC 180
181 LKEKRVSPEK LEELSRYRNK VLEYSKCITA LRSSAYEKDK RYEMLVQKFK RLQKCLKKSE 240
241 NDDDRMSNGG SDCSAASTVS LDTITEDFEE VFAKDIETDY QALYRENAEL QRALNELQLN 300
301 TSDLSEESFL RDQISFANST IEQQQLVIDA TQDMMSQTAQ LKSTIAGQQE HIRSLETTVD 360
361 DLKQQIASQT ERNDVLEFQV LEMEENQKQQ DTIAAKQSTF ESEKTQLLKE LDEVKKAHAN 420
421 QLKSIALIKK ELEDHKAAAP SVKPTLSESE QVDALQQKFE KAEAANVSIN EELRSASKEL 480
481 GKIRFQMQGK ESDLSRERKM TEALSAQLQS VVSSSQEEAL KKEEELKKLK ATVELQQAEI 540
541 EKMHEDKKEA DQKFKNLEKE YAAYRDEQRP EIKTELERRY EEAKYRLKNA LEKIHDYELL 600
601 YEAAKKSEND GSISKHLEEE LIEVKEFNAH LERQFQSQSD IIEALKKKLL QHRSFCDKIN 660
661 KLSELEDASR IQEELVMFSR ENNCDEGKIA ANIALIIKDI RKNTFYSPLE ISTAHHKIRP 720
721 YSSVDSSAEW QSNSSEGIVS PDEHDE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | hcp2 ip from december | Cheeseman, I.M., et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
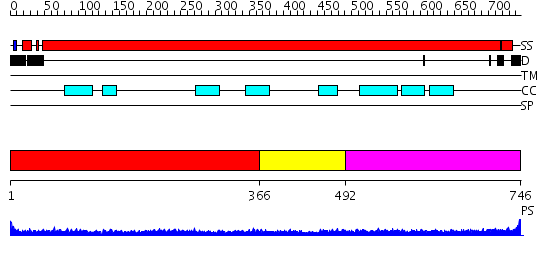
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..365] | 32.522879 | Heavy meromyosin subfragment | |
| 2 | View Details | [366..491] | 1.89 | Ribonuclease domain of colicin E3; Colicin E3 translocation domain; Colicin E3 receptor domain | |
| 3 | View Details | [492..746] | 10.522879 | Tropomyosin |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)