













| General Information: |
|
| Name(s) found: |
CE22611
[WormBase]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 732 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDTPLGRLIN FLVEWTKNVV EPLVGGRIFN RFAELDGAGL SHIYYYIRFH EHIAPNIAKW 60
61 DNFNVTTRIR MMFALSGEVP DDFLALIKLT GTVQLDAKRR ISKYASNDGN LKLEGCHGCT 120
121 ARSKNERGKD PAEITRFMNF LVEKAKVSTD PMFASVVFTE FRKSEEDGLA YSTYCRKFYN 180
181 YLAPRMDQFV NYSIEERVRL MFGLGGEVTE DFLKLCRKEG VVELDKNNRI CEYISYDGTL 240
241 KLGAAHGLSA NMKRRSLLIA KDPSDLPRLM NFLVEKTKDA TEPMVSIAEL SEFRRRERSE 300
301 LTEQAYYGKF HRQLAPKMGQ IVNYSIEERV RVMFGFAGEV TDDFLKQIQT IGTVHLDEKK 360
361 RICKYATHDG KLKLEGDHSH TARMLRRGAY YRRLKTGRKS ISKFSGMEHS RSPKRARISE 420
421 RDDDDEYFGV VNEGESHNRD VSHHNLNIDY LQANNQPIPE DLPYEIDDDE MEHVGAVNYD 480
481 PEHRHFDFDD SHIGHLQNDE EMMNNQSIPE DPSYKSDEVE YNGSLPGLPK PEPIDFDFYQ 540
541 IYNRDVSHSL NLNNLRENKE VSWNPSISEI KVDFTFFERQ QFSVPTNNGA TSAESKIISL 600
601 HDFLKSLIQF ICFLESSELT EIKQKIKESI RTDNDKFLQI TDICTVLQAF FFGISRKFRL 660
661 AAAASNSPAM KMKDFLVKFK LFLLGLDSSE LLELQQKVQE NIDEQTELAE KVLPISDVRQ 720
721 SLQNLLSTIS QY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
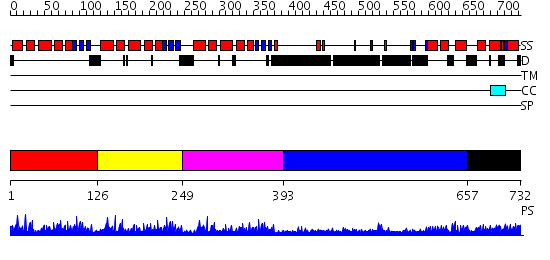
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..125] | 61.657577 | No description for PF04435.9 was found. No confident structure predictions are available. | |
| 2 | View Details | [126..248] | 65.221849 | No description for PF04435.9 was found. No confident structure predictions are available. | |
| 3 | View Details | [249..392] | 68.173925 | No description for PF04435.9 was found. No confident structure predictions are available. | |
| 4 | View Details | [393..656] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [657..732] | 2.030993 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)