













| General Information: |
|
| Name(s) found: |
CE20865
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 740 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
hermaphrodite genitalia development
[IMP nematode larval development [IMP morphogenesis of an epithelium [IMP positive regulation of growth rate [IMP gamete generation [IMP growth [IMP |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNKKKKVSLF GEDDAEEQQN GAGWEINKGY AQKYDNWRRL EEMQKIKDKY GDIDDDDSSS 60
61 ESEPEWTEGH EEAFLKTLGA LKSGDSSCFE NGKGFFKDVQ DAPSKPNNNP KKSKSEKSKK 120
121 KDEKMTIKDY ERKLVLEKGG QIDESDEEAE YNKTQQRGYY EEQEEIRKSL SAAINADDGA 180
181 DDDLLTERKK TSKEQEDEDE DFYQWMKSED GKADKKKAKE LKHLKKFWKD DKNMDESDKF 240
241 LRDYLLNKDY EPGENEENPT YEDIVALEED EKDLDSNRKY EHKFNFRFED PDQEFIKQYP 300
301 RTVAESMRSE DSSRKDKRHE REERKKREKA EKKKELAELK KMKRSEIEQK LGKLQKAAGI 360
361 HIPLTLDELN ADFDPKEFDK KMQSIFNDEY YGVEENIQEN EDEKPIFSDM EDSDYEDYDN 420
421 LDVDELKREE QEADDDDEED EESVDDEDEQ EVPKSKKSAM KNGKFDMVAA AQKAMSKDKN 480
481 DSRRKSKRNA LKDALARKKP LFNPKEKTFE EYFNEYYALD YEDIIGDTPT RFKYRDVESN 540
541 DFGLSTDEIL EADERQLNAW ASLKKVTAYR TSQEEFFDRK AYQRKAEDVN KKKRILSIDF 600
601 GGKKSLKRKM EDEQMEKEMA EMNDQNDNTE EDAEKKKKKK KRGKKKVKLT ETENPEENMT 660
661 EAIENEVEEV DKQEQLPVPQ PEEPEAAHEA PKEEKKKTRK RNKNKNNVIA EKFGQGMTDS 720
721 RVKAYGLNPS RLKKGLVYGQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
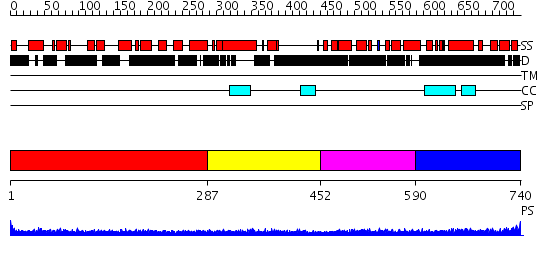
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..286] | 22.0 | Heavy meromyosin subfragment | |
| 2 | View Details | [287..451] | 5.045757 | No description for 2i1jA was found. | |
| 3 | View Details | [452..589] | 62.200659 | No description for PF05178.3 was found. No confident structure predictions are available. | |
| 4 | View Details | [590..740] | 1.08 | THE BLUETONGUE VIRUS (BTV) CORE BINDS DSRNA |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)