













| General Information: |
|
| Name(s) found: |
CE18159
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 659 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
embryonic development ending in birth or egg hatching
[IMP protein modification process [IEA |
| Molecular Function: |
protein-L-isoaspartate (D-aspartate) O-methyltransferase activity
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGNSESQNDD LIDFLVKNDT IRRRNIERAF RLVDRSDFLP ISERKFTRLP SLTSTEPGGP 60
61 FYPGALRVGA IDIYAKLFDY LDLRKGHSFL HIGTGSGYLS TIAGILLGET GINHGIELYE 120
121 NLVTYSETCI DQWITTPEAS SVGWARPELK VCDITDVKFL EAHQNRYDRI FVGFVADDSQ 180
181 LLKRMLGMLN VGGQLVMPRI QGVCRYRRVS KHRTVYDVPG RMTFSPMIRL PKEERISTPT 240
241 PFESGPRTLT YLARQALRRD IRRHAYNPRV NRQICGSIMV MKGGNTYMDH VQRHQAEQFE 300
301 ELNRRRQLFD PRPIDQLTPA DNQLTDQRTR NHLDDVMDQY LGRNNNPREQ ENNEQNIRNT 360
361 IADHVRRWNE TRTDVLQEML NWREMQGFTA PRAHLDGANL RNTAPATENR AVQNLLIFFT 420
421 LRLVFFRFLK ILEFNNFFRL KLRIKTIFNF LKIKISNCVD SEIFHFYFRN VAPAYVLEWT 480
481 INHDRPRHHR ILSSIARPET PNTPRMEIVM IPNINDPSTT GLVERPNGDL SEDEQELSDA 540
541 LEEPQESPST STESQVSEDE EPCCSTDPAS TTSRRRRIRS PKSESAKKPR KSSSPPLDDY 600
601 LKEMAVRQNH EQRAIHLRQR RNLEIAGADG VYQRIIDELP LPPTLKDYVR LVQHEVVLK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
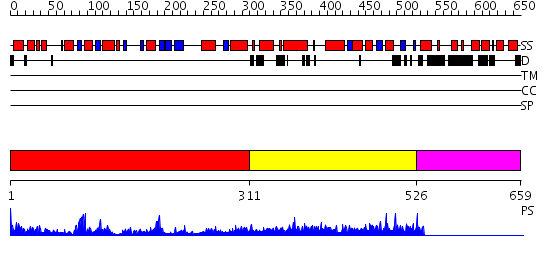
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..310] | 32.522879 | Structure of the cyclopropane synthase MmaA2 from Mycobacterium tuberculosis | |
| 2 | View Details | [311..525] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [526..659] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.80 |
Source: Reynolds et al. (2008)