













| General Information: |
|
| Name(s) found: |
egg-5 /
CE18142
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 753 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
nematode larval development
[IMP embryonic development ending in birth or egg hatching [IMP protein amino acid dephosphorylation [IEA dephosphorylation [IEA reproduction [IMP growth [IMP |
| Molecular Function: |
protein tyrosine phosphatase activity
[IEA phosphatase activity [IEA cytoskeletal protein binding [IEA protein tyrosine/serine/threonine phosphatase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALNSEVMFR EQINAMRSQA GRKRATSLQS FCSGNTDDSS ADSTDNMDMM VDYPQQKGVS 60
61 CMRARFNSES TLSKSFRKKV KKLAQKDRRS KERLNGNSEE DAIEVPRGAP STYAAPSKLR 120
121 KSKALDCLVS EKPKDEGRSE DSGHGADIEM AKGHFNDVRM KVFAARTAMQ VEPALVMKTR 180
181 KALEMKNAVL ENHQSPGAFS LHAAYKIAAS AESRVGSITP CNKKVTKEAM ANLIRSSYDD 240
241 TEITQELLFS SKFDSKWKGR YTDIYMRRDE NGKKPKRPVN GQGWVMPLKS ICEKFGINST 300
301 FFTNHRIDLK SARDQVLLMR LLSHDQTSTW ISDIHPEAVK NETLAEYLLR ELDASTMQKR 360
361 VQAFKANVLA DRDRVRVAGQ FYNNIRIGKR MFGAARKAKY LSTIIGGMER RFEILENSVN 420
421 HIPFTHSASD NNQEKCRNPR VHCRDSTRIA LQFPRGQYLG DFIHANRISG KPLFNEFIMT 480
481 QAPMKNTVDD FWRMVWQEEV PYIVMLTSRK EPERCEYYWP KSPSDPAVTV PGGLRIENFG 540
541 VYQAPDPLFR VTHLRLIGPD REERHVEHWQ GDVNNSSNMY SPLNILRLLR NASKPVVIHD 600
601 HLGVSRAACL VAAEIAICSL LRGPTYKYPV QRAVQFLRQR RPFSIETPMQ YIFVHRLVAF 660
661 FFRDVIGSAK ELDVDYERWL QERSERMFLD DLAAPIPGYR LLSPRADPDI VRMVGRPERP 720
721 NYRREAPDCV GEMPNKVAAV DGILSPAKSV FEF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
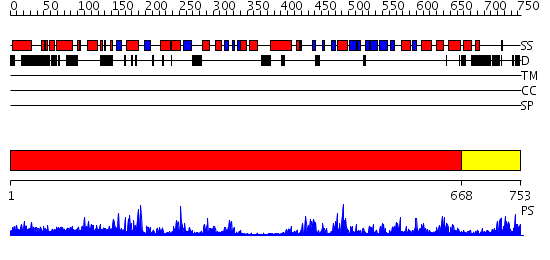
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..667] | 131.0 | Crystal structure of the phosphatase domains of human PTP SIGMA | |
| 2 | View Details | [668..753] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)