













| General Information: |
|
| Name(s) found: |
CE18098
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 782 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
collagen and cuticulin-based cuticle development
[IMP]
reproduction [IMP] |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEQRDMKTVF LLDRSAKFVE NCNETFDVTI REGPKQKKLQ FEKTIWTWCL EGIFEMHRIL 60
61 SDVYPRGTLQ LRFALADFMG KMLDTQWTDG FLTREELGEL IETVSRPSEA NTDITPIGGL 120
121 TMAIEALAVQ SPEQRNYNYD VRYSANKRTA QNSEVIRVTR ELRGLPPLPT VQNAGNLIMY 180
181 TRLNTQEEME QLQTEIVELV VSRNKIADAP SNKTFCPITS LRLFIVNYYA TGDECTVKTH 240
241 SLQAHPDLPL LKIWVISRKA ADMCDAIHSL LVDAFDLGST TVTKIPMKED NRGSSNYDVE 300
301 LFHSGKVHAM LKENNLIDTV SKKGDSDKGV TYDTMRLTWT TAPKTKWSLF PYHGAAVPCT 360
361 TAQAYSRPSA CLTQFVRDGR CVMLDSEKTT ELGMNMHEKL VSHLLIANRG RIFIQEIDFM 420
421 QKKADRIAGK LKRPRVLHAP EDPINMNQLR HTYKQLELRF VAKNRMDDNL TEKETEKWTG 480
481 LKNSDLKKRF QRISKNIPCF AADTFIFNEK VTPKLEPLIS LITKTTLSEN DVDKCKQCIV 540
541 TLYQMKNERQ YVISPESDIK ISSVRNLKDP DEQLRVTFVE LAKHIMKYVT FSEKHKEIYK 600
601 TYMTTLNVDK LLEVDLDDDE AVDKMFVKFS FFGKSDTASK TDEDEDSSDD EDEEILKSPA 660
661 LKKLFGYVDA RDERKYEELY GQGFKKKKPE VEEKPIVMAK EIKVDIFSNM CDMLEVKESK 720
721 IRREFVGRET HGNIAPLYPQ LAEKLERPAS PAATERGTER VRNTAERGGS GTPPPGGRRN 780
781 FQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
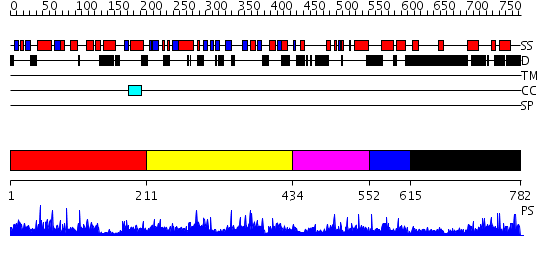
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..210] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [211..433] | 1.012938 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [434..551] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [552..614] | 1.016984 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [615..782] | 1.014986 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)