













| General Information: |
|
| Name(s) found: |
lin-15A /
CE15383
[WormBase]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 719 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
negative regulation of vulval development
[IGI regulation of cell fate specification [IMP nematode larval development [IMP growth [IMP |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLAPAAPAKD VVSADEKEEI IAKRKFRMKN VDAMRMSSLA NDRMAFNKKC NALAMKFVKS 60
61 AGIGTDALQL TCFQELVRHF NPIAAVVVGV KREPNSNVQA EKKTIPKVKT IQTPTQSMES 120
121 VRLLQEKKAS ATEEQSAESA SIMKHFANTI PNSTPTQSVK DVLTAAASKG QFKSSAEIFS 180
181 HFPSEPSPSK PRATREGSQP SDYTYCTYLT PCILCEKALL MRESIAMTDN EAVKVLMAAV 240
241 MSGHFRMATA EKAIRHERLR MCYDHVDFVY EMMCDAFEAK TESEINEMPP DRLMRGHDIY 300
301 RALKRVGDLH KGKVTSNTPL YSFKNSIKSY YRNHVPRMVN GSLSKPSPKP FSELVALLQS 360
361 VPPSTNLNEL LNHNLSLSDA DKQELIQLIN GKDNRFTSRR RKIEDILDNK FAAAAAKAYR 420
421 DHSEDAPSEP YIPNQSEMQN TVERRKRKLH SPEQDDAGSS SISWNAKKTK TPIDYVHLAT 480
481 RVLEGHSIAD EALLHKSKVS YARNAFGEKP SSPTPPSAPL KFCVVNGKKY LRFENGTGPP 540
541 KVVVQGNVVL RTNTLKDALT TAPRAQNQPS TSTDSSSSSE MEGIRQSFGA PQKEEEEEEL 600
601 VPTLLQNKPT HVESSSPVEK KPPTKTNVEK PAVRLGRMLT TAFGSMSYRT RKSVENKTDL 660
661 LNQPTSASPR RMIKVVRNRN PHLAKQVAAA PSEPKHIPPT HMEKKPEELL MDPKPEPIF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
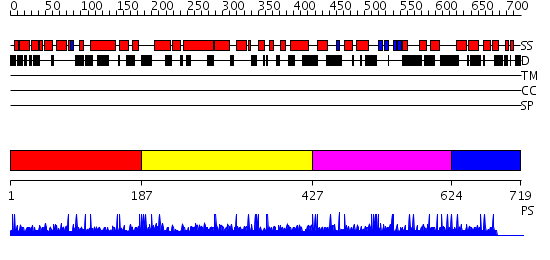
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..186] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [187..426] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [427..623] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [624..719] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)