













| General Information: |
|
| Name(s) found: |
CE09647
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 578 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
cell communication
[IEA positive regulation of growth rate [IMP |
| Molecular Function: |
protein binding
[IEA calcium ion binding [IEA phosphoinositide binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMQSYDYDSD DSQTTSDEHI NSRHSSRSDT ASSSSSFTPQ IIFNPRPHVV KVVKSETGFG 60
61 FNVKGQVSEG GQLRSLNGQL YAPLQHVSAV LRRGAADQAG LRKGDRILEV NGLNVEGSTH 120
121 RKVVDLIKNG GDELTMIVIS VEDPDMDRFD YGEESSMAYG HDYSENRSLP VTIPSYNTVQ 180
181 DSLERYTVFN IHMAGRQLGS RRYSEFVELH LALKKHFYDY CFPQLPGKWP FKLSEQQLDS 240
241 RRRGLEQYLE KICTIRVIAE SELVQKFLME CDPMCEVEIR LMLPDGSPIT IRTRRSITSS 300
301 LFFTSAQRRL KMSREGAAAC SIFELLDNSF ERKVSESESV HELYTHNYSS ASSSCLLLRK 360
361 FIFDIDRERA LCKRDMVFKQ FCFVQAIADL HSGKVVTSRK SYQLRAIQNE ENMDELLEMA 420
421 RELEGYNQVT FPPSVCILRK RRQTVIMVVR FANLLLKCPE DGNSMEIDWS RIKDFRVCDE 480
481 ATAFSFEYIR DEVGDSEQEQ TEPRKTKYVR LETGFAEYMA ECFAEIQFER QAGSDWKRDQ 540
541 NRVAATPEPL PETIEPQPTT HKIQFEKRIL EQHRDCID |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
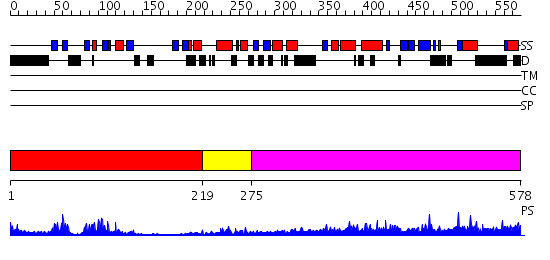
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..218] | 17.522879 | solution structure of the split PH-PDZ Supramodule of alpha-Syntrophin | |
| 2 | View Details | [219..274] | 6.0 | crystal structure of CISK-PX domain | |
| 3 | View Details | [275..578] | 2.76 | Moesin |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)