













| General Information: |
|
| Name(s) found: |
acl-6 /
CE09258
[WormBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 718 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IEA |
| Biological Process: |
metabolic process
[IEA phospholipid biosynthetic process [IEA cellular lipid metabolic process [IEA |
| Molecular Function: |
1-acylglycerol-3-phosphate O-acyltransferase activity
[IEA acyltransferase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MELDVESTSI TQLRSHCETC FPLMEEVVPR RERYVDLLEF SNFNGIPYPV VDPSRKPRRF 60
61 LADFRYSWSI PLIHHYPNVE KDVLSSKRVH RVISKLKEQN DEQKNRAVQF FTEISARLSK 120
121 FICKCCSYVL YKVFRRLMDK LLVCKEEMEV LYEAEQTGIP MVYLPLHRSH LDYLLITWCN 180
181 WHFGLKLPHI ASGDNLNLSG LGWLLRATGA FFIRRRVDPD DERGKDQLYR AILHSYIEQV 240
241 LSKDMPIEFF LEGTRSRFGK ALTPKNGLIS NVVEAVQHGF IKDCYLVPVS YTYDAVVEGI 300
301 FLHELMGIPK VRESVLGVFR GIFSGFSKSK QCGVVRMHYG RPIRLTEYLA TITASLSSNH 360
361 RTRPVRMTKL STSFSYRELV PWHRTHSETV DDRTMIRAIG FHVVYEAQMM CSISPVAVVS 420
421 CLLLAKWRGK VSRSTFERDC EWLCEKIIAE GGDVVGYQSK KTKGSALVKY AFEKLESCVE 480
481 VTDEYVSPKE SHSSFITLAY NKNSVICRFS IKSVIALTIV SRPSGTKLSI DQIVEDALSL 540
541 CDWLQFEFMF CRPCDSLREL VHNVLGQKEW SDPIHGFLRS EIEDDGFLDA GGALNSGTLR 600
601 VRDAKSRETL QFFANLVRPF VQSLYLISSF VVSEKCPTEP TSDNNIIRQL CQQSLAGDID 660
661 LPFAPLLESI NSDSFKNALR VLKDKGLLQR STPNSTARSG NSRLAELISN LERVLEVK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
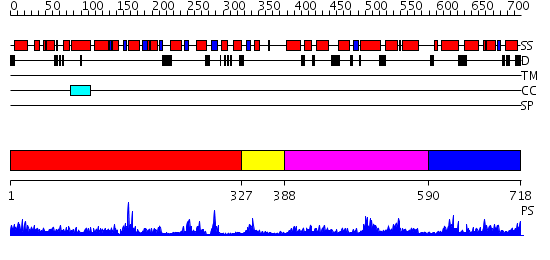
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..326] | 10.09691 | The 1.55 A Crystal Structure of Glycerol-3-Phosphate Acyltransferase | |
| 2 | View Details | [327..387] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [388..589] | 1.14898 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [590..718] | 1.094982 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)