













| General Information: |
|
| Name(s) found: |
obr-4 /
CE08539
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 676 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
steroid metabolic process
[IEA |
| Molecular Function: |
calmodulin binding
[IPI]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEGPLSKWTN MVHGWQYRWF KIENDTLLYY TSREKMLKGQ QRGQIRLTGA VVGIDGENNS 60
61 LFTITVEGKT FHMQGRDLRE RNQWVRILEN SIRASSGYAK NVSVGTTPSQ HFKQRTEEAN 120
121 EFLKTLVEKV KALEQLKSGE RAVDEKKTID SLIAASNNLT NNVKHAIIYL QMAQARLDPN 180
181 MKTEKLMFTH PDLNEKPSSR ETPGTVSDST QTSTDKPISG IQHLDHLKPQ RAESYASSDE 240
241 EEFYDADEEL IDLDNFESEK SGQESGDSNR TSDAAIMSSD EDGYDFGDSN EDFDEIYNNA 300
301 EEHDIGNVQQ DHGSVLMHLL SQVSVGMDLT KVTLPTFILE RRSLLEMYAD FFAHPDLFIA 360
361 TNGNDSAEKR MIAVVRYYLN SFYAARKSGV AKKPYNAILG ETFRCRYSVP NSSLSGKKTE 420
421 SGPWPGSDEK QLTFIAEQVS HHPPISAFYA EMPSEGISFN AHIYTKSSFL GLSIGVASIG 480
481 EGILTLHNYG DEQYTITFPS GYGRSIMSTP WFEFGGKVKV ECEKTGYRAD IEFLTKPFFG 540
541 GKPHRIQGSI CRDGAKKPVL TLRGEWNGTM YAKTQNGEEY VFVDVKAKPE VKKECVPVMQ 600
601 QGKRESRRLW RHVTAALLRN RIQTATTSKR FIEQRQRKEA KERLESGEKW TPLLFDTTDK 660
661 DGEWMYKQKF GVRKAV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
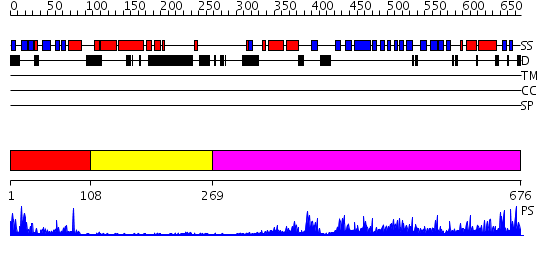
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..107] | 15.39794 | No description for 2d9xA was found. | |
| 2 | View Details | [108..268] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [269..676] | 71.221849 | Structure of yeast oxysterol binding protein Osh4 in complex with 7-hydroxycholesterol |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)