













| General Information: |
|
| Name(s) found: |
CE03847
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 724 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
nematode larval development
[IMP positive regulation of growth rate [IMP embryonic development ending in birth or egg hatching [IMP oviposition [IMP gamete generation [IMP growth [IMP |
| Molecular Function: |
protein binding
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLPNGFSFLN PNLVAAANIQ QVLLNQRFGM PPVGSIAQVP LLQMPTHSVV APHVAAPTRP 60
61 SPMLVPPGMG IDESHSSPSV ESDWSVHTNE KGTPYYHNRV TKQTSWIKPD VLKTPLERST 120
121 SGQPQQGQWK EFMSDDGKPY YYNTLTKKTQ WVKPDGEEIT KGEQKPAAKA ATVDTVALAA 180
181 AVQQKKAESD LDKAMKATLA SMPNVPLPSE KKEEESVNDE VELKKRQSER FRELLRDKYN 240
241 DGKITTNCNW DQAVKWIQND PRFRILNKVS EKKQLFNAWK VQRGKEERDE KRLAIKKSKE 300
301 DLEKFLQEHP KMKESLKYQK ASDIFSKEPL WIAVNDEDRK EIFRDCIDFV ARRDKEKKEE 360
361 DRKRDIAAFS HVLQSMEQIT YKTTWAQAQR ILYENPQFAE RKDLHFMDKE DALTVFEDHI 420
421 KQAEKEHDEE KEQEEKRLRR QQRKVREEYR LLLESLHKRG ELTSMSLWTS LFPIISTDTR 480
481 FELMLFQPGS SPLDLFKFFV EDLKEQYTED RRLIKEILTE KGCQVIATTE YREFSDWVVS 540
541 HEKGGKVDHG NMKLCYNSLI EKAESKAKDE EKESLRRKRR LESEFRNLLK EHNVDKDSEW 600
601 TVIKPKIEKD KAYLAMENDD ERETAFNHYK NGTSGTTAGS EILEKKKKKK DKKKKNKRSD 660
661 NNSESEGEIR EKEKKKKKKH SKEDRMDDEE RGKKSKKSRK RSPSRSESPR HSSEKRKRRE 720
721 SEAD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
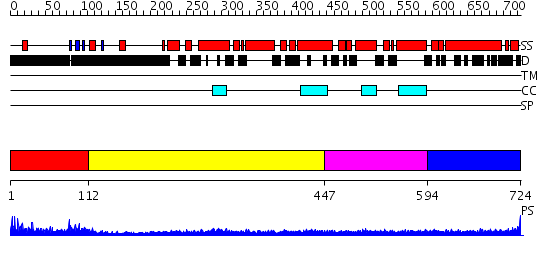
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..111] | 2.17 | No description for 2dk7A was found. | |
| 2 | View Details | [112..446] | 12.09691 | Heavy meromyosin subfragment | |
| 3 | View Details | [447..593] | 5.0 | Tropomyosin | |
| 4 | View Details | [594..724] | 4.69897 | No description for 2i1jA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)